Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
Results for meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2
Z-value: 1.30
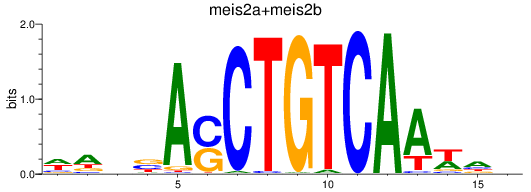
Transcription factors associated with meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meis2b
|
ENSDARG00000077840 | Meis homeobox 2a |
|
meis2a
|
ENSDARG00000098240 | Meis homeobox 2a |
|
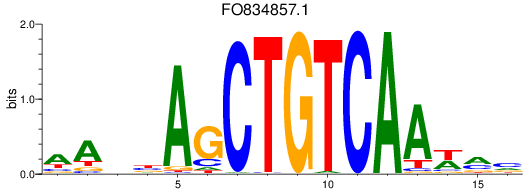
FO834857.1
|
ENSDARG00000112895 | homeobox protein TGIF2-like |
|
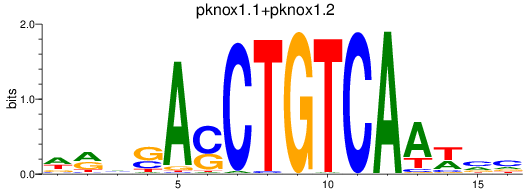
pknox1.1
|
ENSDARG00000018765 | pbx/knotted 1 homeobox 1.1 |
|
pknox1.2
|
ENSDARG00000036542 | pbx/knotted 1 homeobox 1.2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pknox1.2 | dr11_v1_chr1_-_46981134_46981134 | 0.34 | 8.8e-04 | Click! |
| meis2b | dr11_v1_chr20_-_10118818_10118818 | -0.28 | 5.9e-03 | Click! |
| pknox1.1 | dr11_v1_chr9_+_19529951_19529951 | 0.22 | 3.4e-02 | Click! |
| FO834857.1 | dr11_v1_chr6_-_1820606_1820606 | 0.07 | 5.3e-01 | Click! |
| meis2a | dr11_v1_chr17_+_52823015_52823138 | -0.06 | 5.5e-01 | Click! |
Activity profile of meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2 motif
Sorted Z-values of meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_58332000 | 17.04 |
ENSDART00000145234
|
ggt1l2.1
|
gamma-glutamyltransferase 1 like 2.1 |
| chr3_-_36612877 | 13.88 |
ENSDART00000167164
|
si:dkeyp-72e1.7
|
si:dkeyp-72e1.7 |
| chr7_+_14291323 | 11.82 |
ENSDART00000053521
|
rhcga
|
Rh family, C glycoprotein a |
| chr21_-_38153824 | 11.27 |
ENSDART00000151226
|
klf5l
|
Kruppel-like factor 5 like |
| chr19_-_5369486 | 10.64 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr5_-_12587053 | 8.77 |
ENSDART00000162780
|
vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr19_+_7567763 | 8.70 |
ENSDART00000140411
|
s100a11
|
S100 calcium binding protein A11 |
| chr8_+_34731982 | 8.60 |
ENSDART00000066050
|
hpdb
|
4-hydroxyphenylpyruvate dioxygenase b |
| chr21_+_45627775 | 8.58 |
ENSDART00000185466
|
irf1b
|
interferon regulatory factor 1b |
| chr7_+_51103416 | 8.37 |
ENSDART00000174236
|
col4a5
|
collagen, type IV, alpha 5 (Alport syndrome) |
| chr4_+_7841627 | 7.86 |
ENSDART00000037997
|
ucmaa
|
upper zone of growth plate and cartilage matrix associated a |
| chr22_+_19188809 | 7.42 |
ENSDART00000134791
ENSDART00000133682 |
si:dkey-21e2.8
|
si:dkey-21e2.8 |
| chr20_-_27733683 | 7.28 |
ENSDART00000103317
ENSDART00000138139 |
zgc:153157
|
zgc:153157 |
| chr9_-_48281941 | 7.05 |
ENSDART00000099787
|
klhl41a
|
kelch-like family member 41a |
| chr15_+_27364394 | 6.89 |
ENSDART00000122101
|
tbx2b
|
T-box 2b |
| chr5_+_72145468 | 6.81 |
ENSDART00000148626
|
abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr12_-_48168135 | 6.54 |
ENSDART00000186624
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr4_+_73085993 | 6.50 |
ENSDART00000165749
|
si:ch73-170d6.2
|
si:ch73-170d6.2 |
| chr9_-_31135901 | 6.48 |
ENSDART00000113027
ENSDART00000193210 ENSDART00000128896 |
si:ch211-184m13.4
|
si:ch211-184m13.4 |
| chr1_-_8653385 | 6.26 |
ENSDART00000193041
|
actb1
|
actin, beta 1 |
| chr24_+_10413484 | 6.25 |
ENSDART00000111014
|
myca
|
MYC proto-oncogene, bHLH transcription factor a |
| chr24_-_40667800 | 6.08 |
ENSDART00000169315
|
smyhc1
|
slow myosin heavy chain 1 |
| chr17_-_23673864 | 6.06 |
ENSDART00000104738
ENSDART00000128958 |
ptena
|
phosphatase and tensin homolog A |
| chr4_+_22613625 | 6.02 |
ENSDART00000131161
|
sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr16_+_23397785 | 6.01 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr1_-_19402802 | 5.90 |
ENSDART00000135552
|
rbm47
|
RNA binding motif protein 47 |
| chr2_-_11027258 | 5.85 |
ENSDART00000081072
ENSDART00000193824 ENSDART00000187036 ENSDART00000097741 |
ssbp3a
|
single stranded DNA binding protein 3a |
| chr22_-_6194517 | 5.81 |
ENSDART00000134757
ENSDART00000181598 ENSDART00000129829 |
si:rp71-36a1.5
|
si:rp71-36a1.5 |
| chr5_+_30741730 | 5.79 |
ENSDART00000098246
ENSDART00000186992 ENSDART00000182533 |
ftr83
|
finTRIM family, member 83 |
| chr12_-_44307963 | 5.78 |
ENSDART00000161009
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr23_-_9768700 | 5.53 |
ENSDART00000045126
|
lama5
|
laminin, alpha 5 |
| chr20_+_29634653 | 5.51 |
ENSDART00000101556
|
asap2b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2b |
| chr10_+_35417099 | 5.50 |
ENSDART00000063398
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr5_-_323712 | 5.40 |
ENSDART00000188793
|
HOOK3
|
hook microtubule tethering protein 3 |
| chr8_-_32385989 | 5.31 |
ENSDART00000143716
ENSDART00000098850 |
lipg
|
lipase, endothelial |
| chr14_+_22022441 | 5.27 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr5_+_22307605 | 5.25 |
ENSDART00000138154
|
arhgap20b
|
Rho GTPase activating protein 20b |
| chr6_+_23931236 | 5.19 |
ENSDART00000166079
|
gadd45ab
|
growth arrest and DNA-damage-inducible, alpha, b |
| chr16_+_23398369 | 5.15 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr24_+_42148140 | 5.13 |
ENSDART00000010658
|
serpinb1l3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 3 |
| chr12_-_43982343 | 5.10 |
ENSDART00000161539
|
CR385054.1
|
|
| chr6_+_49052741 | 5.03 |
ENSDART00000011876
|
sycp1
|
synaptonemal complex protein 1 |
| chr13_-_42749916 | 4.87 |
ENSDART00000140019
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr18_+_29402623 | 4.85 |
ENSDART00000014703
|
mafa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
| chr19_-_38611814 | 4.85 |
ENSDART00000151958
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr11_-_39202915 | 4.84 |
ENSDART00000105133
|
wnt4a
|
wingless-type MMTV integration site family, member 4a |
| chr1_-_58000438 | 4.63 |
ENSDART00000163761
|
si:ch211-114l13.9
|
si:ch211-114l13.9 |
| chr22_-_17677947 | 4.43 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr12_+_18358082 | 4.42 |
ENSDART00000123315
|
si:dkey-7e14.3
|
si:dkey-7e14.3 |
| chr17_-_24890843 | 4.35 |
ENSDART00000184984
ENSDART00000135569 ENSDART00000193661 |
gale
|
UDP-galactose-4-epimerase |
| chr21_-_27338639 | 4.30 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr3_-_50147160 | 4.28 |
ENSDART00000191341
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr1_+_57187794 | 4.28 |
ENSDART00000152485
|
si:dkey-27j5.9
|
si:dkey-27j5.9 |
| chr15_-_19724932 | 4.25 |
ENSDART00000152345
|
sytl2b
|
synaptotagmin-like 2b |
| chr7_+_6879534 | 4.24 |
ENSDART00000157731
|
zgc:175248
|
zgc:175248 |
| chr1_+_14253118 | 4.16 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr13_+_45524475 | 4.13 |
ENSDART00000074567
ENSDART00000019113 |
maco1b
|
macoilin 1b |
| chr7_+_17816006 | 4.12 |
ENSDART00000080834
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr15_-_28587147 | 4.11 |
ENSDART00000156049
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr23_-_19103855 | 4.11 |
ENSDART00000016901
|
pard6b
|
par-6 partitioning defective 6 homolog beta (C. elegans) |
| chr1_+_58150000 | 4.06 |
ENSDART00000137836
|
si:ch211-15j1.3
|
si:ch211-15j1.3 |
| chr5_-_56513825 | 4.05 |
ENSDART00000024207
|
tbx2a
|
T-box 2a |
| chr17_-_7792376 | 4.04 |
ENSDART00000064655
|
zbtb2a
|
zinc finger and BTB domain containing 2a |
| chr18_+_44673990 | 4.04 |
ENSDART00000018625
|
napab
|
N-ethylmaleimide-sensitive factor attachment protein, alpha b |
| chr3_+_22273123 | 4.03 |
ENSDART00000044157
|
scn4ab
|
sodium channel, voltage-gated, type IV, alpha, b |
| chr18_+_29403017 | 4.02 |
ENSDART00000176966
|
mafa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
| chr4_+_12931763 | 4.00 |
ENSDART00000016382
|
wif1
|
wnt inhibitory factor 1 |
| chr16_+_33144306 | 3.98 |
ENSDART00000101953
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr4_-_685412 | 3.96 |
ENSDART00000168167
|
rtcb
|
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr8_+_25616946 | 3.96 |
ENSDART00000133983
|
slc38a5a
|
solute carrier family 38, member 5a |
| chr13_+_46803979 | 3.92 |
ENSDART00000159260
|
CU695232.1
|
|
| chr13_+_18321140 | 3.91 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr1_+_2301961 | 3.89 |
ENSDART00000108919
ENSDART00000143361 ENSDART00000142944 |
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr11_+_27364338 | 3.88 |
ENSDART00000186759
|
fbln2
|
fibulin 2 |
| chr8_+_23639124 | 3.87 |
ENSDART00000083108
|
nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr3_-_1400309 | 3.86 |
ENSDART00000159893
|
wbp11
|
WW domain binding protein 11 |
| chr21_-_41025340 | 3.81 |
ENSDART00000148231
|
plac8l1
|
PLAC8-like 1 |
| chr13_+_30696286 | 3.80 |
ENSDART00000192411
|
cxcl18a.1
|
chemokine (C-X-C motif) ligand 18a, duplicate 1 |
| chr14_-_34605607 | 3.74 |
ENSDART00000191608
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr20_+_32152355 | 3.69 |
ENSDART00000152904
ENSDART00000139507 |
sesn1
|
sestrin 1 |
| chr13_-_7031033 | 3.68 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr14_+_35369979 | 3.67 |
ENSDART00000144702
ENSDART00000169712 |
clint1a
|
clathrin interactor 1a |
| chr12_-_46176115 | 3.62 |
ENSDART00000152848
|
si:ch211-226h7.8
|
si:ch211-226h7.8 |
| chr12_-_36511939 | 3.61 |
ENSDART00000153759
|
unc13d
|
unc-13 homolog D (C. elegans) |
| chr12_-_43693192 | 3.60 |
ENSDART00000172284
|
CU914622.1
|
|
| chr6_-_10809546 | 3.60 |
ENSDART00000151661
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
| chr16_-_52646789 | 3.59 |
ENSDART00000035761
|
ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr20_+_21391181 | 3.58 |
ENSDART00000185158
ENSDART00000049586 ENSDART00000024922 |
jag2b
|
jagged 2b |
| chr23_+_20408227 | 3.52 |
ENSDART00000134727
|
si:rp71-17i16.4
|
si:rp71-17i16.4 |
| chr7_+_17816470 | 3.51 |
ENSDART00000173807
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr3_+_17030665 | 3.48 |
ENSDART00000159849
ENSDART00000174491 ENSDART00000104519 ENSDART00000080854 |
stat3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr5_-_71722257 | 3.45 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr9_+_27876146 | 3.45 |
ENSDART00000133997
|
armc8
|
armadillo repeat containing 8 |
| chr1_+_1805294 | 3.41 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 3 |
| chr1_-_30473422 | 3.39 |
ENSDART00000164202
|
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr1_-_10821584 | 3.36 |
ENSDART00000167452
ENSDART00000162137 |
crfb15
|
cytokine receptor family member B15 |
| chr16_+_1383914 | 3.35 |
ENSDART00000185089
|
cers2b
|
ceramide synthase 2b |
| chr6_+_46320693 | 3.31 |
ENSDART00000128621
ENSDART00000155917 |
si:dkeyp-67f1.2
|
si:dkeyp-67f1.2 |
| chr2_+_394166 | 3.28 |
ENSDART00000155733
|
mylk4a
|
myosin light chain kinase family, member 4a |
| chr16_+_33144112 | 3.25 |
ENSDART00000183149
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr12_+_33320884 | 3.14 |
ENSDART00000188988
|
csnk1db
|
casein kinase 1, delta b |
| chr5_-_37103487 | 3.14 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr3_-_55099272 | 3.12 |
ENSDART00000130869
|
hbba1
|
hemoglobin, beta adult 1 |
| chr14_+_29769336 | 3.12 |
ENSDART00000105898
|
si:dkey-34l15.1
|
si:dkey-34l15.1 |
| chr7_+_65673885 | 3.07 |
ENSDART00000169182
|
parvab
|
parvin, alpha b |
| chr20_-_54564018 | 3.06 |
ENSDART00000099832
|
zgc:153012
|
zgc:153012 |
| chr3_-_55104310 | 3.05 |
ENSDART00000101713
|
hbba1
|
hemoglobin, beta adult 1 |
| chr11_+_27274355 | 3.03 |
ENSDART00000113707
|
fbln2
|
fibulin 2 |
| chr4_+_76775837 | 3.03 |
ENSDART00000174167
|
ms4a17a.10
|
membrane-spanning 4-domains, subfamily A, member 17A.10 |
| chr13_+_35765317 | 3.01 |
ENSDART00000100156
ENSDART00000167650 |
agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr2_+_55365727 | 2.99 |
ENSDART00000162943
|
FP245456.1
|
|
| chr23_-_20309505 | 2.98 |
ENSDART00000130856
|
lamb2l
|
laminin, beta 2-like |
| chr5_-_41124241 | 2.97 |
ENSDART00000083561
|
mtmr12
|
myotubularin related protein 12 |
| chr1_-_11075403 | 2.91 |
ENSDART00000102903
ENSDART00000170290 |
dmd
|
dystrophin |
| chr18_-_6943577 | 2.91 |
ENSDART00000132399
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr11_+_45436703 | 2.91 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr15_-_14212777 | 2.90 |
ENSDART00000165572
|
CR925813.1
|
|
| chr19_+_7575141 | 2.88 |
ENSDART00000051528
|
s100u
|
S100 calcium binding protein U |
| chr11_+_27364059 | 2.85 |
ENSDART00000172883
|
fbln2
|
fibulin 2 |
| chr19_-_35035857 | 2.84 |
ENSDART00000103253
|
bmp8a
|
bone morphogenetic protein 8a |
| chr7_+_17816623 | 2.83 |
ENSDART00000173963
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr15_-_28587490 | 2.83 |
ENSDART00000186196
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr3_+_17616201 | 2.82 |
ENSDART00000156775
|
rab5c
|
RAB5C, member RAS oncogene family |
| chr8_+_7033049 | 2.74 |
ENSDART00000064172
ENSDART00000134440 |
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr5_-_35200590 | 2.73 |
ENSDART00000051271
|
fcho2
|
FCH domain only 2 |
| chr10_-_8294965 | 2.72 |
ENSDART00000167380
|
plpp1a
|
phospholipid phosphatase 1a |
| chr15_+_36445350 | 2.71 |
ENSDART00000154552
|
si:dkey-262k9.2
|
si:dkey-262k9.2 |
| chr19_-_6134802 | 2.71 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr11_+_25459697 | 2.70 |
ENSDART00000161481
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr18_+_35128685 | 2.69 |
ENSDART00000151579
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr15_-_35252522 | 2.65 |
ENSDART00000144153
ENSDART00000059195 |
mff
|
mitochondrial fission factor |
| chr4_+_76692834 | 2.65 |
ENSDART00000064315
ENSDART00000137653 |
ms4a17a.1
|
membrane-spanning 4-domains, subfamily A, member 17A.1 |
| chr9_-_52490579 | 2.62 |
ENSDART00000161667
|
smarcal1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr5_+_44346691 | 2.61 |
ENSDART00000034523
|
tars
|
threonyl-tRNA synthetase |
| chr5_-_24127310 | 2.61 |
ENSDART00000182700
ENSDART00000154313 |
capga
|
capping protein (actin filament), gelsolin-like a |
| chr9_+_33267211 | 2.53 |
ENSDART00000025635
|
usp9
|
ubiquitin specific peptidase 9 |
| chr20_-_20270191 | 2.52 |
ENSDART00000009356
|
ppp2r5ea
|
protein phosphatase 2, regulatory subunit B', epsilon isoform a |
| chr5_-_39805620 | 2.51 |
ENSDART00000137801
|
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr19_+_7575341 | 2.50 |
ENSDART00000134271
|
s100u
|
S100 calcium binding protein U |
| chr23_-_38054 | 2.49 |
ENSDART00000170393
|
CABZ01074076.1
|
|
| chr11_-_29916607 | 2.48 |
ENSDART00000138912
ENSDART00000079152 |
vegfd
|
vascular endothelial growth factor D |
| chr20_+_1329509 | 2.47 |
ENSDART00000017791
ENSDART00000136669 |
tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr9_-_22821901 | 2.47 |
ENSDART00000101711
|
neb
|
nebulin |
| chr13_-_7233811 | 2.40 |
ENSDART00000162026
|
ninl
|
ninein-like |
| chr9_-_7539297 | 2.40 |
ENSDART00000081550
ENSDART00000081553 |
desma
|
desmin a |
| chr5_-_37900350 | 2.36 |
ENSDART00000084839
ENSDART00000084841 ENSDART00000133437 |
tmprss13b
|
transmembrane protease, serine 13b |
| chr23_-_36592436 | 2.36 |
ENSDART00000168246
|
spryd3
|
SPRY domain containing 3 |
| chr14_+_48862987 | 2.35 |
ENSDART00000167810
|
zgc:154054
|
zgc:154054 |
| chr7_+_34794829 | 2.35 |
ENSDART00000009698
ENSDART00000075089 ENSDART00000173456 |
esrp2
|
epithelial splicing regulatory protein 2 |
| chr7_-_71331690 | 2.30 |
ENSDART00000149682
|
dhx15
|
DEAH (Asp-Glu-Ala-His) box helicase 15 |
| chr13_-_12581388 | 2.28 |
ENSDART00000079655
|
enpep
|
glutamyl aminopeptidase |
| chr19_+_19772765 | 2.27 |
ENSDART00000182028
ENSDART00000161019 |
hoxa3a
|
homeobox A3a |
| chr14_-_25956804 | 2.27 |
ENSDART00000135627
ENSDART00000146022 ENSDART00000039660 |
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr10_+_26667475 | 2.26 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr10_-_8295294 | 2.26 |
ENSDART00000075412
ENSDART00000163803 |
plpp1a
|
phospholipid phosphatase 1a |
| chr13_-_15700060 | 2.20 |
ENSDART00000170689
ENSDART00000010986 ENSDART00000101741 ENSDART00000139124 |
ckba
|
creatine kinase, brain a |
| chr6_-_49510553 | 2.19 |
ENSDART00000166238
|
rplp2
|
ribosomal protein, large P2 |
| chr6_+_58915889 | 2.19 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr18_+_24562188 | 2.15 |
ENSDART00000099463
|
lysmd4
|
LysM, putative peptidoglycan-binding, domain containing 4 |
| chr25_-_21822426 | 2.12 |
ENSDART00000151993
|
zgc:158222
|
zgc:158222 |
| chr14_-_34605804 | 2.09 |
ENSDART00000144547
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr8_+_23709280 | 2.07 |
ENSDART00000185842
|
ppardb
|
peroxisome proliferator-activated receptor delta b |
| chr11_+_5880562 | 2.07 |
ENSDART00000129663
ENSDART00000130768 ENSDART00000160909 |
dazap1
|
DAZ associated protein 1 |
| chr25_+_5068442 | 2.03 |
ENSDART00000097522
|
parvg
|
parvin, gamma |
| chr20_+_26690036 | 2.02 |
ENSDART00000103232
|
foxf2b
|
forkhead box F2b |
| chr12_-_18898413 | 2.01 |
ENSDART00000181281
ENSDART00000121866 |
desi1b
|
desumoylating isopeptidase 1b |
| chr10_+_20590190 | 2.01 |
ENSDART00000131819
|
letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr2_+_36103309 | 1.99 |
ENSDART00000190038
|
CT867973.1
|
|
| chr5_+_27432958 | 1.98 |
ENSDART00000124705
|
histh1l
|
histone H1 like |
| chr2_+_3696038 | 1.97 |
ENSDART00000150499
ENSDART00000136906 ENSDART00000164152 ENSDART00000128514 ENSDART00000108964 |
egfra
|
epidermal growth factor receptor a (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) |
| chr1_+_41478345 | 1.95 |
ENSDART00000134626
|
dok1a
|
docking protein 1a |
| chr6_-_42003780 | 1.95 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr7_-_71331499 | 1.94 |
ENSDART00000081245
|
dhx15
|
DEAH (Asp-Glu-Ala-His) box helicase 15 |
| chr19_-_12212692 | 1.93 |
ENSDART00000142077
ENSDART00000151599 ENSDART00000140834 ENSDART00000078781 |
znf706
|
zinc finger protein 706 |
| chr23_+_6586467 | 1.92 |
ENSDART00000081763
ENSDART00000121480 |
rbm38
|
RNA binding motif protein 38 |
| chr23_-_20258490 | 1.91 |
ENSDART00000147326
|
lamb2
|
laminin, beta 2 (laminin S) |
| chr1_+_54865552 | 1.88 |
ENSDART00000145381
|
si:ch211-196h16.5
|
si:ch211-196h16.5 |
| chr3_+_29941777 | 1.88 |
ENSDART00000113889
|
ifi35
|
interferon-induced protein 35 |
| chr7_-_56766100 | 1.88 |
ENSDART00000189934
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr21_+_45758498 | 1.85 |
ENSDART00000162548
|
h2afy
|
H2A histone family, member Y |
| chr2_-_45510699 | 1.85 |
ENSDART00000024034
ENSDART00000145634 |
gpsm2
|
G protein signaling modulator 2 |
| chr3_-_50146854 | 1.84 |
ENSDART00000157047
|
si:dkey-120c6.5
|
si:dkey-120c6.5 |
| chr22_-_37834312 | 1.84 |
ENSDART00000076128
|
ppp1r2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr3_+_53317040 | 1.84 |
ENSDART00000011780
|
xab2
|
XPA binding protein 2 |
| chr7_+_21768452 | 1.83 |
ENSDART00000127719
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr3_+_13190012 | 1.82 |
ENSDART00000179747
ENSDART00000109876 ENSDART00000124824 ENSDART00000130261 |
sun1
|
Sad1 and UNC84 domain containing 1 |
| chr19_-_81477 | 1.80 |
ENSDART00000159815
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr22_-_17458070 | 1.79 |
ENSDART00000139658
|
si:ch211-197g15.10
|
si:ch211-197g15.10 |
| chr1_-_52437056 | 1.78 |
ENSDART00000138337
|
si:ch211-217k17.12
|
si:ch211-217k17.12 |
| chr3_+_24275766 | 1.74 |
ENSDART00000055607
|
pdgfbb
|
platelet-derived growth factor beta polypeptide b |
| chr19_-_2861444 | 1.74 |
ENSDART00000169053
|
clec3bb
|
C-type lectin domain family 3, member Bb |
| chr9_-_22822084 | 1.67 |
ENSDART00000142020
|
neb
|
nebulin |
| chr20_+_27712714 | 1.67 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr5_-_23795688 | 1.65 |
ENSDART00000099084
|
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr12_-_48467733 | 1.64 |
ENSDART00000153126
ENSDART00000152895 ENSDART00000014190 |
sec31b
|
SEC31 homolog B, COPII coat complex component |
| chr23_-_23401305 | 1.62 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr8_-_17184482 | 1.62 |
ENSDART00000025803
|
pola2
|
polymerase (DNA directed), alpha 2 |
| chr16_+_5251768 | 1.61 |
ENSDART00000144558
|
plecb
|
plectin b |
| chr11_+_6136220 | 1.61 |
ENSDART00000082223
|
tax1bp3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr11_+_44356504 | 1.60 |
ENSDART00000160678
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
Network of associatons between targets according to the STRING database.
First level regulatory network of meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0072025 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 2.2 | 17.9 | GO:0031179 | peptide modification(GO:0031179) |
| 1.7 | 5.0 | GO:0051026 | meiotic DNA repair synthesis(GO:0000711) chiasma assembly(GO:0051026) |
| 1.7 | 8.4 | GO:0007412 | axon target recognition(GO:0007412) |
| 1.5 | 11.8 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 1.5 | 5.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 1.4 | 4.2 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 1.2 | 6.1 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 1.2 | 3.5 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 1.1 | 3.3 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 1.0 | 4.0 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 1.0 | 4.0 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 1.0 | 6.9 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.9 | 4.3 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.8 | 4.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.8 | 5.3 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.7 | 8.6 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.7 | 4.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.7 | 2.7 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.7 | 2.0 | GO:1903392 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.6 | 2.4 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.6 | 3.6 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.6 | 5.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.5 | 2.2 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.5 | 1.6 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.5 | 3.7 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.5 | 7.9 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.5 | 3.0 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.5 | 5.5 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.5 | 3.4 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.5 | 2.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.4 | 4.4 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.4 | 1.8 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.4 | 1.3 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.4 | 7.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.4 | 2.0 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.4 | 1.2 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.4 | 4.0 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.4 | 1.6 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.4 | 2.6 | GO:0048478 | embryonic body morphogenesis(GO:0010172) replication fork protection(GO:0048478) |
| 0.4 | 3.2 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.3 | 1.0 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.3 | 1.3 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.3 | 2.3 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.3 | 1.0 | GO:0001783 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.3 | 1.8 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.3 | 1.5 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.3 | 1.2 | GO:0060468 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.3 | 1.2 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.3 | 6.6 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.3 | 3.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.3 | 1.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.3 | 1.0 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.3 | 3.9 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.3 | 1.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.3 | 0.8 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.2 | 0.7 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 2.5 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.2 | 5.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 1.9 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) muscle cell cellular homeostasis(GO:0046716) caveola assembly(GO:0070836) |
| 0.2 | 5.5 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.2 | 4.0 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.2 | 3.8 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 1.3 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.2 | 1.4 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.2 | 1.6 | GO:0042026 | protein refolding(GO:0042026) |
| 0.2 | 2.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 0.6 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.2 | 5.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 3.4 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 1.8 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.2 | 2.5 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.2 | 4.3 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.2 | 0.8 | GO:0060760 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.2 | 2.2 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 0.8 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.2 | 0.8 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.2 | 0.8 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) mitochondrial respiratory chain complex III assembly(GO:0034551) protein insertion into mitochondrial membrane(GO:0051204) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 0.7 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.2 | 1.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 0.5 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 1.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 1.0 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 1.2 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 15.6 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 0.4 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.1 | 1.4 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 0.7 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 3.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.7 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.8 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 2.3 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 2.5 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.6 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.1 | 0.5 | GO:0006297 | leading strand elongation(GO:0006272) nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 2.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.1 | 9.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 2.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 5.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 6.3 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of chemotaxis(GO:0050922) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 4.0 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.1 | 0.5 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 10.1 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.1 | 2.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 3.9 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 1.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 1.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.4 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 0.5 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.1 | 0.5 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.8 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 1.4 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 1.7 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.7 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.6 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 1.8 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.1 | 0.7 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.1 | 0.7 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 0.8 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 4.5 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 0.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 1.0 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 2.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 1.0 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.4 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 4.1 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.1 | 1.4 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.1 | 3.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.5 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 1.8 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 17.2 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 0.4 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.1 | 0.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.6 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 5.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 1.3 | GO:0071166 | ribonucleoprotein complex localization(GO:0071166) |
| 0.1 | 1.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.9 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 0.7 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 7.0 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 0.6 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.2 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 2.5 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 4.7 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.1 | 0.3 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 1.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 5.4 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 6.0 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.1 | 0.4 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 2.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.7 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 0.3 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 3.8 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.1 | 0.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 3.1 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.9 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 0.0 | 1.1 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 1.6 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 1.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.6 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 1.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 2.6 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.7 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 1.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 1.2 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 1.5 | GO:0010906 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 4.1 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 4.6 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 2.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.3 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 9.1 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 1.7 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.0 | 0.3 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.5 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.3 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.8 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.3 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.6 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.4 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.4 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 1.6 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 2.1 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.3 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 1.2 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.3 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.3 | GO:0009251 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.9 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.2 | GO:0048016 | calcineurin-NFAT signaling cascade(GO:0033173) inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 6.5 | GO:0001525 | angiogenesis(GO:0001525) |
| 0.0 | 0.1 | GO:0032329 | serine transport(GO:0032329) |
| 0.0 | 0.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.3 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 7.1 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 0.2 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.0 | 0.1 | GO:0034694 | response to prostaglandin(GO:0034694) cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.8 | GO:0032869 | response to insulin(GO:0032868) cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 0.3 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.7 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.6 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.2 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.0 | GO:0030162 | regulation of proteolysis(GO:0030162) |
| 0.0 | 0.5 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.5 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.4 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.0 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 1.3 | 6.3 | GO:0097433 | dense body(GO:0097433) |
| 1.0 | 4.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.7 | 3.4 | GO:0034657 | GID complex(GO:0034657) |
| 0.6 | 4.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.5 | 1.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.5 | 10.8 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.5 | 3.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.5 | 2.7 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.4 | 6.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.4 | 5.8 | GO:0002102 | podosome(GO:0002102) |
| 0.4 | 1.2 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.4 | 1.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.4 | 2.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.4 | 1.5 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.3 | 1.6 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.3 | 1.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.3 | 1.4 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.3 | 7.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 1.8 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.2 | 3.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 1.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 4.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.2 | 4.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 2.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.2 | 1.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.2 | 6.9 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.2 | 6.1 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 3.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.2 | 16.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 3.6 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 3.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 2.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.0 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.9 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 0.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 1.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.8 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.1 | 8.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.6 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 1.3 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 0.4 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.1 | 2.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 4.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 2.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 2.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.8 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.7 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 4.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 5.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 1.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 7.9 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 3.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 10.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 1.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 6.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.8 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.4 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.1 | 1.1 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 2.8 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 0.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 5.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 8.3 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 5.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 4.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 2.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 12.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 55.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.3 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 1.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.7 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 3.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 3.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 1.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.0 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 11.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 11.9 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 1.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 1.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.6 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 8.6 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.0 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 4.4 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 1.7 | GO:0099503 | secretory vesicle(GO:0099503) |
| 0.0 | 0.4 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 2.0 | 17.9 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 1.7 | 5.0 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 1.5 | 6.1 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 1.4 | 4.3 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 1.3 | 6.3 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 1.0 | 4.2 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 1.0 | 28.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.9 | 2.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.8 | 2.5 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.8 | 4.0 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.7 | 2.7 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.7 | 4.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.7 | 2.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.5 | 3.7 | GO:0070728 | leucine binding(GO:0070728) |
| 0.5 | 1.9 | GO:0071253 | connexin binding(GO:0071253) |
| 0.5 | 5.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.5 | 11.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.4 | 2.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.4 | 6.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.4 | 1.5 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 3.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 4.0 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.3 | 2.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 4.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.3 | 7.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.3 | 0.8 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.3 | 3.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 1.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 0.7 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 5.2 | GO:0005344 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.2 | 11.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 1.2 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.2 | 1.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 4.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 2.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 3.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 3.0 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 1.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 24.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 1.8 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 2.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 1.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 2.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.2 | 4.0 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 1.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 1.5 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 0.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 4.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 4.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 2.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 1.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.4 | GO:0031834 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.1 | 3.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 3.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.0 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 2.5 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 9.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.7 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 3.0 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 0.6 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.7 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.1 | 1.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 3.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 2.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 1.4 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.3 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 5.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.4 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 1.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 7.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.2 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.1 | 15.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 4.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 1.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.3 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 1.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.3 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.1 | 1.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.1 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 2.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 1.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 3.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 2.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 6.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 1.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 4.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 1.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 2.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.6 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 2.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 4.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 0.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.3 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 5.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 1.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 3.3 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 1.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 2.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 3.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 4.2 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 21.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 4.2 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 1.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 6.0 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 9.8 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.5 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 4.0 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 4.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.1 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 1.0 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 1.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 2.7 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 7.1 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.5 | 3.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.4 | 3.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.3 | 13.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 6.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 4.4 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.2 | 4.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 11.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 5.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 3.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 2.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 17.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 0.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 1.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 13.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 5.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 2.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 3.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | PID BCR 5PATHWAY | BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.5 | 4.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.4 | 4.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.4 | 3.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 14.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 7.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 1.5 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.2 | 2.9 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.2 | 0.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 6.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 6.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 2.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.2 | 3.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 5.9 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 1.2 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 3.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 7.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 0.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 2.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 0.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 2.4 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 0.6 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 4.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.5 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 0.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 2.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.7 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.9 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 2.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 2.7 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.0 | 0.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 2.6 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 0.4 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |