Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
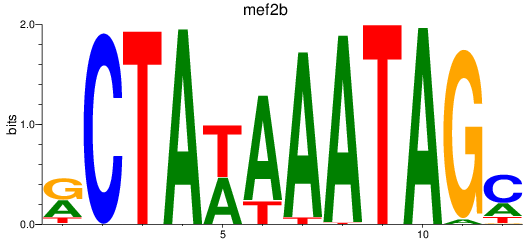
Results for mef2b
Z-value: 1.39
Transcription factors associated with mef2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mef2b
|
ENSDARG00000093170 | myocyte enhancer factor 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mef2b | dr11_v1_chr22_+_18187857_18187857 | 0.33 | 1.0e-03 | Click! |
Activity profile of mef2b motif
Sorted Z-values of mef2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_61205711 | 32.68 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr8_+_22930627 | 20.82 |
ENSDART00000187860
|
sypa
|
synaptophysin a |
| chr11_+_11201096 | 20.57 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr5_-_31926906 | 18.64 |
ENSDART00000187340
|
ssh1b
|
slingshot protein phosphatase 1b |
| chr5_+_64368770 | 17.57 |
ENSDART00000162246
|
plpp7
|
phospholipid phosphatase 7 (inactive) |
| chr3_-_32818607 | 17.16 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr1_+_45080897 | 17.00 |
ENSDART00000129819
|
si:ch211-151p13.8
|
si:ch211-151p13.8 |
| chr6_-_40722480 | 15.65 |
ENSDART00000188187
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr25_+_29160102 | 15.13 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr23_+_22658700 | 14.85 |
ENSDART00000192248
|
eno1a
|
enolase 1a, (alpha) |
| chr19_-_31035155 | 14.56 |
ENSDART00000161882
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr9_-_33107237 | 14.27 |
ENSDART00000013918
|
casq2
|
calsequestrin 2 |
| chr25_-_13381854 | 13.80 |
ENSDART00000164621
ENSDART00000169129 |
ndrg4
|
NDRG family member 4 |
| chr16_+_29663809 | 13.45 |
ENSDART00000191336
|
tmod4
|
tropomodulin 4 (muscle) |
| chr6_-_40722200 | 13.34 |
ENSDART00000035101
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr5_+_51597677 | 13.32 |
ENSDART00000048210
ENSDART00000184797 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr14_+_24215046 | 13.25 |
ENSDART00000079215
|
stc2a
|
stanniocalcin 2a |
| chr6_-_14139503 | 13.11 |
ENSDART00000089577
|
cacnb4b
|
calcium channel, voltage-dependent, beta 4b subunit |
| chr7_-_23745984 | 12.98 |
ENSDART00000048050
|
ITGB1BP2
|
zgc:92429 |
| chr10_-_22845485 | 12.60 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr5_-_72125551 | 12.22 |
ENSDART00000149412
|
smyd1a
|
SET and MYND domain containing 1a |
| chr24_+_25471196 | 12.06 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr23_-_39849155 | 11.95 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr17_-_12336987 | 11.72 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr19_-_31035325 | 11.71 |
ENSDART00000147504
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr1_-_25936677 | 11.48 |
ENSDART00000146488
ENSDART00000136321 |
myoz2b
|
myozenin 2b |
| chr10_-_8033468 | 10.89 |
ENSDART00000140476
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr10_-_24371312 | 10.76 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr1_+_8601935 | 10.25 |
ENSDART00000152367
|
si:ch211-160d14.6
|
si:ch211-160d14.6 |
| chr6_-_35472923 | 9.85 |
ENSDART00000185907
|
rgs8
|
regulator of G protein signaling 8 |
| chr10_-_8032885 | 9.84 |
ENSDART00000188619
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr6_-_10780698 | 9.40 |
ENSDART00000151714
|
gpr155b
|
G protein-coupled receptor 155b |
| chr25_-_29363934 | 9.13 |
ENSDART00000166889
|
nptna
|
neuroplastin a |
| chr20_-_26001288 | 8.98 |
ENSDART00000136518
ENSDART00000063177 |
capn3b
|
calpain 3b |
| chr7_-_52334840 | 8.80 |
ENSDART00000174173
|
CR938716.1
|
|
| chr18_+_7264961 | 8.78 |
ENSDART00000188461
|
CABZ01015105.1
|
|
| chr20_+_41549200 | 8.72 |
ENSDART00000135715
|
fam184a
|
family with sequence similarity 184, member A |
| chr1_-_21483832 | 8.63 |
ENSDART00000102790
|
glrba
|
glycine receptor, beta a |
| chr6_+_36942966 | 8.42 |
ENSDART00000028895
|
negr1
|
neuronal growth regulator 1 |
| chr20_-_26042070 | 7.99 |
ENSDART00000140255
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr14_-_32405387 | 7.88 |
ENSDART00000184647
|
fgf13a
|
fibroblast growth factor 13a |
| chr10_+_37145007 | 7.79 |
ENSDART00000131777
|
cuedc1a
|
CUE domain containing 1a |
| chr4_-_23908802 | 7.69 |
ENSDART00000138873
|
celf2
|
cugbp, Elav-like family member 2 |
| chr6_-_42003780 | 7.58 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr17_-_14671098 | 7.39 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr13_+_9432501 | 7.39 |
ENSDART00000058064
|
zgc:123321
|
zgc:123321 |
| chr21_+_11684830 | 7.38 |
ENSDART00000147473
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr12_+_6002715 | 7.30 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr23_-_32157865 | 7.24 |
ENSDART00000000876
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr5_-_51819027 | 6.84 |
ENSDART00000164267
|
homer1b
|
homer scaffolding protein 1b |
| chr21_-_27185915 | 6.54 |
ENSDART00000135052
|
slc8a4a
|
solute carrier family 8 (sodium/calcium exchanger), member 4a |
| chr5_-_10946232 | 6.51 |
ENSDART00000163139
ENSDART00000031265 |
rtn4r
|
reticulon 4 receptor |
| chr19_+_41169996 | 6.11 |
ENSDART00000048438
|
asb4
|
ankyrin repeat and SOCS box containing 4 |
| chr13_-_27767330 | 6.09 |
ENSDART00000131631
ENSDART00000112553 ENSDART00000189911 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr13_+_1944451 | 6.06 |
ENSDART00000125914
|
hmgcll1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
| chr25_+_3677650 | 5.98 |
ENSDART00000154348
|
prnprs3
|
prion protein, related sequence 3 |
| chr12_-_26430507 | 5.95 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr21_+_11685009 | 5.88 |
ENSDART00000014668
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr12_+_41697664 | 5.69 |
ENSDART00000162302
|
bnip3
|
BCL2 interacting protein 3 |
| chr1_-_6494384 | 5.55 |
ENSDART00000109356
|
klf7a
|
Kruppel-like factor 7a |
| chr3_+_32425202 | 5.54 |
ENSDART00000156464
|
prr12b
|
proline rich 12b |
| chr19_+_24394560 | 5.48 |
ENSDART00000142506
|
si:dkey-81h8.1
|
si:dkey-81h8.1 |
| chr10_-_2522588 | 5.28 |
ENSDART00000081926
|
CU856539.1
|
|
| chr21_-_12272543 | 5.24 |
ENSDART00000081510
ENSDART00000151297 |
celf4
|
CUGBP, Elav-like family member 4 |
| chr1_-_17569793 | 5.08 |
ENSDART00000125125
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr9_+_31795343 | 4.87 |
ENSDART00000139584
|
itgbl1
|
integrin, beta-like 1 |
| chr6_-_46403475 | 4.86 |
ENSDART00000154148
|
camk1a
|
calcium/calmodulin-dependent protein kinase Ia |
| chr21_+_19070921 | 4.75 |
ENSDART00000029874
|
nkx6.1
|
NK6 homeobox 1 |
| chr2_+_19777146 | 4.74 |
ENSDART00000038648
|
ptbp2b
|
polypyrimidine tract binding protein 2b |
| chr10_-_41450367 | 4.70 |
ENSDART00000122682
ENSDART00000189549 |
cabp1b
|
calcium binding protein 1b |
| chr10_+_29698467 | 4.68 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr12_+_42400277 | 4.67 |
ENSDART00000166590
|
si:ch211-221j21.3
|
si:ch211-221j21.3 |
| chr17_-_5769196 | 4.58 |
ENSDART00000113885
|
si:dkey-100n19.2
|
si:dkey-100n19.2 |
| chr21_+_43559123 | 4.50 |
ENSDART00000151212
|
gpr185a
|
G protein-coupled receptor 185 a |
| chr17_-_10025234 | 4.43 |
ENSDART00000008355
|
cfl2
|
cofilin 2 (muscle) |
| chr7_-_27686021 | 4.36 |
ENSDART00000079112
ENSDART00000100989 |
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr11_-_18253111 | 4.34 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr7_+_21859337 | 4.27 |
ENSDART00000159626
|
si:dkey-85k7.7
|
si:dkey-85k7.7 |
| chr2_-_6182098 | 4.24 |
ENSDART00000156167
|
si:ch73-182a11.2
|
si:ch73-182a11.2 |
| chr9_-_48937240 | 4.22 |
ENSDART00000075627
|
cers6
|
ceramide synthase 6 |
| chr16_+_25245857 | 4.21 |
ENSDART00000155220
|
klhl38b
|
kelch-like family member 38b |
| chr19_+_30990129 | 4.19 |
ENSDART00000052169
ENSDART00000193376 |
sync
|
syncoilin, intermediate filament protein |
| chr5_+_51227147 | 4.17 |
ENSDART00000083340
|
ubac1
|
UBA domain containing 1 |
| chr21_+_39432248 | 4.16 |
ENSDART00000179938
|
pafah1b1b
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1b |
| chr7_-_49646251 | 4.08 |
ENSDART00000193674
|
hrasb
|
-Ha-ras Harvey rat sarcoma viral oncogene homolog b |
| chr10_-_2527342 | 4.01 |
ENSDART00000184168
|
CU856539.1
|
|
| chr12_+_8373525 | 3.94 |
ENSDART00000152180
|
arid5b
|
AT-rich interaction domain 5B |
| chr24_-_28333029 | 3.93 |
ENSDART00000149015
ENSDART00000129174 |
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr25_-_27722614 | 3.87 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr15_-_14552101 | 3.82 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr7_+_39006837 | 3.73 |
ENSDART00000173735
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr14_+_30285613 | 3.55 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr23_+_6077503 | 3.54 |
ENSDART00000081714
ENSDART00000139834 |
mybpha
|
myosin binding protein Ha |
| chr8_-_26388090 | 3.53 |
ENSDART00000147912
|
si:dkey-20d21.12
|
si:dkey-20d21.12 |
| chr19_-_7321221 | 3.50 |
ENSDART00000092375
|
oxr1b
|
oxidation resistance 1b |
| chr13_+_11439486 | 3.46 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr4_+_5180650 | 3.43 |
ENSDART00000067390
|
fgf6b
|
fibroblast growth factor 6b |
| chr9_-_48281941 | 3.39 |
ENSDART00000099787
|
klhl41a
|
kelch-like family member 41a |
| chr12_-_34035364 | 3.38 |
ENSDART00000087065
|
timp2a
|
TIMP metallopeptidase inhibitor 2a |
| chr20_+_34455645 | 3.38 |
ENSDART00000135789
|
mettl11b
|
methyltransferase like 11B |
| chr12_+_27022517 | 3.35 |
ENSDART00000152975
|
msl1b
|
male-specific lethal 1 homolog b (Drosophila) |
| chr2_+_1202347 | 3.16 |
ENSDART00000075837
|
CABZ01084566.1
|
|
| chr18_+_26719787 | 3.15 |
ENSDART00000141672
|
alpk3a
|
alpha-kinase 3a |
| chr25_-_29988352 | 3.13 |
ENSDART00000067059
|
fam19a5b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5b |
| chr9_-_23894392 | 3.09 |
ENSDART00000133417
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
| chr11_-_18601955 | 3.00 |
ENSDART00000180565
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr9_+_32178374 | 2.92 |
ENSDART00000078576
|
coq10b
|
coenzyme Q10B |
| chr4_-_5018705 | 2.92 |
ENSDART00000154025
|
strip2
|
striatin interacting protein 2 |
| chr5_-_50781623 | 2.91 |
ENSDART00000114950
|
zgc:194908
|
zgc:194908 |
| chr23_+_20110086 | 2.86 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr23_+_17417539 | 2.86 |
ENSDART00000182605
|
BX649300.2
|
|
| chr24_-_12745222 | 2.84 |
ENSDART00000151836
|
si:ch211-196f5.9
|
si:ch211-196f5.9 |
| chr1_-_17570013 | 2.79 |
ENSDART00000146946
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr12_+_34119439 | 2.77 |
ENSDART00000032821
|
cyth1b
|
cytohesin 1b |
| chr7_-_31938938 | 2.72 |
ENSDART00000132353
|
bdnf
|
brain-derived neurotrophic factor |
| chr19_-_41069573 | 2.63 |
ENSDART00000111982
ENSDART00000193142 |
sgce
|
sarcoglycan, epsilon |
| chr13_-_36581875 | 2.57 |
ENSDART00000113204
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr7_+_31379528 | 2.42 |
ENSDART00000187370
|
ctdspl2b
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2b |
| chr4_-_4261673 | 2.36 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr4_+_13449775 | 2.35 |
ENSDART00000172552
|
dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr18_+_3169579 | 2.33 |
ENSDART00000164724
ENSDART00000186340 ENSDART00000181247 ENSDART00000168056 |
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr23_-_3721444 | 2.28 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr20_+_45620076 | 2.21 |
ENSDART00000113806
|
hnrnpa0l
|
heterogeneous nuclear ribonucleoprotein A0, like |
| chr14_-_24332786 | 2.18 |
ENSDART00000173164
|
fam13b
|
family with sequence similarity 13, member B |
| chr5_+_61657702 | 2.12 |
ENSDART00000134387
ENSDART00000171248 |
ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr5_-_31875645 | 2.03 |
ENSDART00000098160
|
tmem119b
|
transmembrane protein 119b |
| chr12_+_30563550 | 1.97 |
ENSDART00000126064
|
si:ch211-28p3.4
|
si:ch211-28p3.4 |
| chr21_-_25669820 | 1.96 |
ENSDART00000148236
|
tmem179b
|
transmembrane protein 179B |
| chr5_+_64842730 | 1.95 |
ENSDART00000144732
|
lrrc8ab
|
leucine rich repeat containing 8 VRAC subunit Ab |
| chr8_+_17987215 | 1.93 |
ENSDART00000113605
|
lrriq3
|
leucine-rich repeats and IQ motif containing 3 |
| chr25_-_19486399 | 1.88 |
ENSDART00000155076
ENSDART00000156016 |
zgc:193812
|
zgc:193812 |
| chr18_+_17959681 | 1.75 |
ENSDART00000142700
|
znf423
|
zinc finger protein 423 |
| chr23_-_36670369 | 1.68 |
ENSDART00000006881
|
zbtb39
|
zinc finger and BTB domain containing 39 |
| chr9_+_29040425 | 1.66 |
ENSDART00000150201
|
MRAS
|
si:ch73-116o1.2 |
| chr25_-_27722309 | 1.66 |
ENSDART00000148121
|
zgc:153935
|
zgc:153935 |
| chr6_-_46768040 | 1.57 |
ENSDART00000154071
|
igfn1.2
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 2 |
| chr23_-_7826849 | 1.55 |
ENSDART00000157612
|
myt1b
|
myelin transcription factor 1b |
| chr22_+_2861734 | 1.52 |
ENSDART00000140578
|
si:dkey-20i20.2
|
si:dkey-20i20.2 |
| chr17_-_50234004 | 1.49 |
ENSDART00000058706
|
fosaa
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Aa |
| chr9_-_48184823 | 1.43 |
ENSDART00000180264
|
klhl23
|
kelch-like family member 23 |
| chr19_+_44039849 | 1.42 |
ENSDART00000086040
|
lrrc14b
|
leucine rich repeat containing 14B |
| chr4_+_14343706 | 1.37 |
ENSDART00000142845
|
prl2
|
prolactin 2 |
| chr5_-_18911114 | 1.33 |
ENSDART00000014434
|
bri3bp
|
bri3 binding protein |
| chr5_+_61658282 | 1.32 |
ENSDART00000188878
|
ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr3_+_26135502 | 1.24 |
ENSDART00000146979
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr5_-_30151815 | 1.18 |
ENSDART00000156048
|
zbtb44
|
zinc finger and BTB domain containing 44 |
| chr24_+_8736497 | 1.17 |
ENSDART00000181904
|
tmem14ca
|
transmembrane protein 14Ca |
| chr9_+_21793565 | 1.17 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr8_+_7097929 | 1.15 |
ENSDART00000188955
ENSDART00000184772 ENSDART00000109581 |
abtb1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr7_+_39679944 | 1.15 |
ENSDART00000133420
|
tbc1d14
|
TBC1 domain family, member 14 |
| chr23_-_38497705 | 1.13 |
ENSDART00000109493
|
tshz2
|
teashirt zinc finger homeobox 2 |
| chr13_+_42011287 | 1.04 |
ENSDART00000131147
|
cyp1b1
|
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chr6_+_14301214 | 0.94 |
ENSDART00000129491
|
tmem182b
|
transmembrane protein 182b |
| chr13_-_24825691 | 0.92 |
ENSDART00000142745
|
slka
|
STE20-like kinase a |
| chr18_+_49248389 | 0.84 |
ENSDART00000059285
ENSDART00000142004 ENSDART00000132751 |
yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr15_-_14755236 | 0.81 |
ENSDART00000166880
ENSDART00000160302 |
p4ha3
|
prolyl 4-hydroxylase, alpha polypeptide III |
| chr2_+_37975026 | 0.77 |
ENSDART00000034802
|
si:rp71-1g18.13
|
si:rp71-1g18.13 |
| chr9_+_32178050 | 0.77 |
ENSDART00000169526
|
coq10b
|
coenzyme Q10B |
| chr23_-_25050329 | 0.72 |
ENSDART00000140216
|
avpr2aa
|
arginine vasopressin receptor 2a, duplicate a |
| chr1_-_45662774 | 0.66 |
ENSDART00000042158
|
serhl
|
serine hydrolase-like |
| chr3_+_17456428 | 0.65 |
ENSDART00000090676
ENSDART00000182082 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr24_+_9178064 | 0.64 |
ENSDART00000142971
|
dlgap1b
|
discs, large (Drosophila) homolog-associated protein 1b |
| chr23_+_23485858 | 0.55 |
ENSDART00000114067
|
agrn
|
agrin |
| chr4_+_77060861 | 0.54 |
ENSDART00000174271
ENSDART00000174393 ENSDART00000150450 |
si:dkey-240n22.8
|
si:dkey-240n22.8 |
| chr23_+_3721042 | 0.53 |
ENSDART00000143323
|
smim29
|
small integral membrane protein 29 |
| chr16_-_30655980 | 0.46 |
ENSDART00000146508
|
ldlrad4b
|
low density lipoprotein receptor class A domain containing 4b |
| chr6_+_46697710 | 0.45 |
ENSDART00000154969
|
tespa1
|
thymocyte expressed, positive selection associated 1 |
| chr9_-_28399071 | 0.45 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr19_-_32518556 | 0.41 |
ENSDART00000103410
|
zbtb8b
|
zinc finger and BTB domain containing 8B |
| chr8_+_44420108 | 0.39 |
ENSDART00000075381
|
CU571323.1
|
|
| chr8_-_53166975 | 0.37 |
ENSDART00000114683
|
rbsn
|
rabenosyn, RAB effector |
| chr10_-_17170086 | 0.36 |
ENSDART00000020122
|
ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
| chr15_+_20543770 | 0.35 |
ENSDART00000092357
|
sgsm2
|
small G protein signaling modulator 2 |
| chr25_+_32530976 | 0.28 |
ENSDART00000156190
ENSDART00000103324 |
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr20_-_24443680 | 0.27 |
ENSDART00000191337
|
CR385053.3
|
|
| chr17_+_1992495 | 0.26 |
ENSDART00000192937
|
CABZ01062996.1
|
|
| chr7_-_7420301 | 0.24 |
ENSDART00000102620
|
six7
|
SIX homeobox 7 |
| chr3_+_31717291 | 0.24 |
ENSDART00000058221
|
dcaf7
|
ddb1 and cul4 associated factor 7 |
| chr5_-_25236340 | 0.23 |
ENSDART00000162774
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr5_+_16580739 | 0.22 |
ENSDART00000135140
|
htr7c
|
5-hydroxytryptamine (serotonin) receptor 7c |
| chr5_+_67390645 | 0.19 |
ENSDART00000014822
|
ebf2
|
early B cell factor 2 |
| chr5_+_67390115 | 0.16 |
ENSDART00000193255
|
ebf2
|
early B cell factor 2 |
| chr9_+_24065855 | 0.16 |
ENSDART00000161468
ENSDART00000171577 ENSDART00000172743 ENSDART00000159324 ENSDART00000079689 ENSDART00000023196 ENSDART00000101577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr16_-_26435431 | 0.12 |
ENSDART00000187526
|
megf8
|
multiple EGF-like-domains 8 |
| chr8_-_17980317 | 0.04 |
ENSDART00000129148
|
tnni3k
|
TNNI3 interacting kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of mef2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 14.3 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 3.0 | 9.0 | GO:0097264 | self proteolysis(GO:0097264) |
| 2.5 | 9.9 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 1.8 | 25.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 1.6 | 8.0 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 1.5 | 12.3 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 1.5 | 6.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 1.5 | 13.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 1.5 | 20.7 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 1.5 | 4.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 1.2 | 12.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 1.2 | 6.0 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 1.1 | 3.4 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 1.0 | 4.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 1.0 | 13.3 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 1.0 | 6.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.9 | 13.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.9 | 4.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.8 | 7.6 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.8 | 12.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.8 | 7.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.7 | 10.3 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.7 | 2.0 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.6 | 5.7 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.6 | 8.6 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.6 | 3.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.6 | 7.9 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.5 | 6.5 | GO:0042044 | fluid transport(GO:0042044) |
| 0.5 | 7.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.5 | 2.3 | GO:1901906 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.4 | 3.9 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.4 | 6.3 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.4 | 2.3 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.3 | 12.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.3 | 3.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.3 | 13.1 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.3 | 9.1 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.3 | 7.9 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.3 | 1.2 | GO:0031446 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.3 | 1.2 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.3 | 6.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 2.9 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.3 | 5.9 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.2 | 2.7 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 6.0 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.2 | 4.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 15.1 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.2 | 15.7 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.2 | 3.7 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 0.5 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.2 | 3.4 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.2 | 10.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.2 | 3.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 4.9 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.2 | 0.5 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 21.9 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 4.4 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 3.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 3.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 1.1 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 1.0 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.7 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.1 | 4.2 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 2.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.4 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 1.4 | GO:0046427 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.1 | 3.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 13.3 | GO:0006874 | cellular calcium ion homeostasis(GO:0006874) |
| 0.0 | 0.8 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 2.4 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 1.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.6 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 4.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 3.5 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 3.3 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 4.7 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 3.0 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 12.1 | GO:0043034 | costamere(GO:0043034) |
| 2.1 | 20.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 2.0 | 14.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 2.0 | 11.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 1.7 | 32.8 | GO:0031430 | M band(GO:0031430) |
| 1.5 | 12.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.0 | 4.2 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.5 | 9.9 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.4 | 3.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.4 | 2.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 8.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.3 | 12.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.3 | 3.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 26.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 1.2 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 16.3 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.2 | 3.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 17.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 4.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 13.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 7.6 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 7.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 3.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 4.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 3.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 5.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 6.8 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.6 | GO:0099572 | postsynaptic specialization(GO:0099572) |
| 0.0 | 4.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 8.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 6.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 8.1 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 11.8 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 1.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 4.9 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 3.1 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.8 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 15.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.9 | 7.6 | GO:0071253 | connexin binding(GO:0071253) |
| 1.5 | 12.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 1.5 | 10.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 1.5 | 6.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 1.5 | 4.4 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 1.4 | 20.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 1.4 | 6.8 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 1.3 | 12.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.3 | 10.3 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 1.1 | 11.5 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 1.1 | 45.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 1.1 | 3.4 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 1.0 | 13.3 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 1.0 | 8.6 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.7 | 7.9 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.7 | 7.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.6 | 13.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.5 | 6.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.5 | 7.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.5 | 2.3 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.4 | 13.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.4 | 7.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.4 | 3.7 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.4 | 6.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 4.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.3 | 7.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 4.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 9.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.3 | 3.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 18.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 0.7 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.2 | 2.7 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.2 | 12.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.2 | 1.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 3.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 3.9 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 6.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 3.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 4.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 2.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 4.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 14.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 4.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 7.7 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 0.7 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.8 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 13.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 53.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 18.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 3.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 3.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.7 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 7.2 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 6.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 4.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.4 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 13.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.4 | 7.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.3 | 7.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 2.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 9.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 5.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 4.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 4.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 2.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.7 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.6 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 1.3 | 13.3 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.3 | 3.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.3 | 2.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 7.2 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.2 | 4.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 4.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 9.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 1.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 6.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |