Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
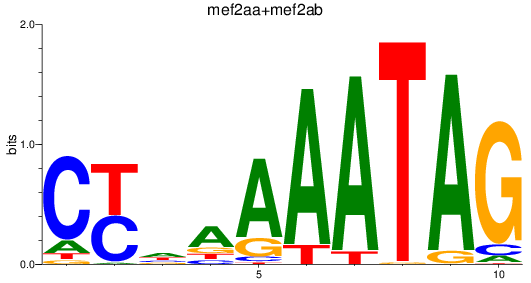
Results for mef2aa+mef2ab
Z-value: 2.89
Transcription factors associated with mef2aa+mef2ab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mef2aa
|
ENSDARG00000031756 | myocyte enhancer factor 2aa |
|
mef2ab
|
ENSDARG00000057527 | myocyte enhancer factor 2ab |
|
mef2ab
|
ENSDARG00000110771 | myocyte enhancer factor 2ab |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mef2aa | dr11_v1_chr18_+_23249519_23249519 | 0.80 | 7.2e-22 | Click! |
| mef2ab | dr11_v1_chr7_+_15677196_15677196 | 0.23 | 2.4e-02 | Click! |
Activity profile of mef2aa+mef2ab motif
Sorted Z-values of mef2aa+mef2ab motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_61205711 | 78.08 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr25_+_31264155 | 68.42 |
ENSDART00000012256
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr3_+_32526263 | 60.37 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr12_-_26064105 | 58.95 |
ENSDART00000168825
|
ldb3b
|
LIM domain binding 3b |
| chr5_-_72125551 | 52.59 |
ENSDART00000149412
|
smyd1a
|
SET and MYND domain containing 1a |
| chr21_+_20383837 | 51.01 |
ENSDART00000026430
|
hspb11
|
heat shock protein, alpha-crystallin-related, b11 |
| chr6_-_18199062 | 49.70 |
ENSDART00000167513
|
ppp1r27b
|
protein phosphatase 1, regulatory subunit 27b |
| chr8_-_11112058 | 47.93 |
ENSDART00000042755
|
ampd1
|
adenosine monophosphate deaminase 1 (isoform M) |
| chr12_-_17712393 | 46.75 |
ENSDART00000143534
ENSDART00000010144 |
pvalb2
|
parvalbumin 2 |
| chr7_+_7048245 | 46.75 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr16_+_29663809 | 44.46 |
ENSDART00000191336
|
tmod4
|
tropomodulin 4 (muscle) |
| chr19_-_7450796 | 42.76 |
ENSDART00000104750
|
mllt11
|
MLLT11, transcription factor 7 cofactor |
| chr12_-_26064480 | 41.96 |
ENSDART00000158215
ENSDART00000171206 ENSDART00000171212 ENSDART00000182956 ENSDART00000186779 |
ldb3b
|
LIM domain binding 3b |
| chr1_+_7546259 | 40.42 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr5_+_51597677 | 40.31 |
ENSDART00000048210
ENSDART00000184797 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr7_+_6969909 | 37.23 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr14_+_49296052 | 35.07 |
ENSDART00000006073
ENSDART00000105346 |
anxa6
|
annexin A6 |
| chr16_+_25245857 | 30.80 |
ENSDART00000155220
|
klhl38b
|
kelch-like family member 38b |
| chr24_+_34089977 | 29.69 |
ENSDART00000157466
|
asb10
|
ankyrin repeat and SOCS box containing 10 |
| chr9_-_42873700 | 29.47 |
ENSDART00000125953
|
ttn.1
|
titin, tandem duplicate 1 |
| chr11_+_11200550 | 29.17 |
ENSDART00000181339
ENSDART00000187116 |
myom2a
|
myomesin 2a |
| chr17_-_5583345 | 28.96 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr6_+_14301214 | 28.60 |
ENSDART00000129491
|
tmem182b
|
transmembrane protein 182b |
| chr20_-_26001288 | 28.28 |
ENSDART00000136518
ENSDART00000063177 |
capn3b
|
calpain 3b |
| chr5_+_64368770 | 27.49 |
ENSDART00000162246
|
plpp7
|
phospholipid phosphatase 7 (inactive) |
| chr13_-_2215213 | 27.32 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr3_-_52614747 | 26.97 |
ENSDART00000154365
|
trim35-13
|
tripartite motif containing 35-13 |
| chr23_+_20110086 | 26.57 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr13_+_22480857 | 26.47 |
ENSDART00000078721
ENSDART00000044719 ENSDART00000130957 ENSDART00000078757 ENSDART00000130424 ENSDART00000078747 |
ldb3a
|
LIM domain binding 3a |
| chr7_-_23745984 | 26.24 |
ENSDART00000048050
|
ITGB1BP2
|
zgc:92429 |
| chr2_-_15324837 | 25.82 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr12_+_6002715 | 25.58 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr1_+_6171585 | 24.88 |
ENSDART00000024358
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr7_+_69841017 | 24.46 |
ENSDART00000169107
|
FO818704.1
|
|
| chr25_+_20077225 | 24.30 |
ENSDART00000136543
|
tnni4b.1
|
troponin I4b, tandem duplicate 1 |
| chr11_-_18253111 | 24.24 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr5_-_57635664 | 24.00 |
ENSDART00000074306
|
hspb2
|
heat shock protein, alpha-crystallin-related, b2 |
| chr18_+_26719787 | 23.87 |
ENSDART00000141672
|
alpk3a
|
alpha-kinase 3a |
| chr15_+_5923851 | 23.47 |
ENSDART00000152520
ENSDART00000145827 ENSDART00000121529 |
sh3bgr
|
SH3 domain binding glutamate-rich protein |
| chr14_+_22113331 | 22.35 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr19_-_33087246 | 20.95 |
ENSDART00000052078
|
klhl38a
|
kelch-like family member 38a |
| chr16_-_43356018 | 20.73 |
ENSDART00000181683
|
FO704821.1
|
|
| chr8_-_11229523 | 19.87 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr23_+_6077503 | 19.80 |
ENSDART00000081714
ENSDART00000139834 |
mybpha
|
myosin binding protein Ha |
| chr19_-_31035155 | 19.25 |
ENSDART00000161882
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr25_+_14087045 | 19.25 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr23_+_24926407 | 19.10 |
ENSDART00000137486
|
klhl21
|
kelch-like family member 21 |
| chr21_-_131236 | 19.10 |
ENSDART00000160005
|
si:ch1073-398f15.1
|
si:ch1073-398f15.1 |
| chr25_-_23745765 | 18.36 |
ENSDART00000089278
|
si:ch211-236l14.4
|
si:ch211-236l14.4 |
| chr16_-_17200120 | 18.22 |
ENSDART00000147739
|
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr19_-_31035325 | 18.09 |
ENSDART00000147504
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr24_+_9298198 | 16.89 |
ENSDART00000165780
|
otud1
|
OTU deubiquitinase 1 |
| chr9_-_23891102 | 16.67 |
ENSDART00000186799
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
| chr17_-_14671098 | 16.63 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr6_-_20875111 | 16.25 |
ENSDART00000115118
ENSDART00000159916 |
tns1a
|
tensin 1a |
| chr21_-_27507786 | 15.75 |
ENSDART00000077657
ENSDART00000191284 ENSDART00000178229 ENSDART00000186153 |
CAPN1
|
si:dkeyp-50d11.2 |
| chr20_+_34455645 | 15.11 |
ENSDART00000135789
|
mettl11b
|
methyltransferase like 11B |
| chr24_+_25471196 | 15.02 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr19_+_30990815 | 14.86 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr8_-_26388090 | 14.78 |
ENSDART00000147912
|
si:dkey-20d21.12
|
si:dkey-20d21.12 |
| chr3_-_32170850 | 14.66 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr1_-_11075403 | 12.64 |
ENSDART00000102903
ENSDART00000170290 |
dmd
|
dystrophin |
| chr5_-_48260145 | 12.42 |
ENSDART00000044083
ENSDART00000163250 ENSDART00000135911 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr18_+_23193567 | 12.31 |
ENSDART00000190072
ENSDART00000171594 ENSDART00000181762 |
mef2aa
|
myocyte enhancer factor 2aa |
| chr6_+_12462079 | 12.20 |
ENSDART00000192029
ENSDART00000065385 |
nr4a2b
|
nuclear receptor subfamily 4, group A, member 2b |
| chr3_+_28953274 | 12.14 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr9_+_54644626 | 11.95 |
ENSDART00000190609
|
egfl6
|
EGF-like-domain, multiple 6 |
| chr23_+_17417539 | 11.55 |
ENSDART00000182605
|
BX649300.2
|
|
| chr23_+_33907899 | 11.53 |
ENSDART00000159445
|
cs
|
citrate synthase |
| chr20_-_36575475 | 11.48 |
ENSDART00000062893
|
enah
|
enabled homolog (Drosophila) |
| chr12_-_4781801 | 11.40 |
ENSDART00000167490
ENSDART00000121718 |
mapta
|
microtubule-associated protein tau a |
| chr18_+_23193820 | 11.28 |
ENSDART00000148106
|
mef2aa
|
myocyte enhancer factor 2aa |
| chr11_-_40030139 | 11.15 |
ENSDART00000021916
|
uts2b
|
urotensin 2, beta |
| chr2_-_23768818 | 10.79 |
ENSDART00000148685
ENSDART00000191167 |
xirp1
|
xin actin binding repeat containing 1 |
| chr15_-_22139566 | 10.72 |
ENSDART00000149017
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr25_+_11389220 | 10.53 |
ENSDART00000187787
|
AGBL1
|
si:ch73-141f14.1 |
| chr7_-_19614916 | 10.45 |
ENSDART00000169029
|
zgc:194655
|
zgc:194655 |
| chrM_+_4993 | 10.35 |
ENSDART00000093600
|
mt-nd2
|
NADH dehydrogenase 2, mitochondrial |
| chr24_+_38155830 | 9.75 |
ENSDART00000152019
|
si:ch211-234p6.5
|
si:ch211-234p6.5 |
| chr11_-_33960318 | 9.53 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr3_-_53114299 | 9.41 |
ENSDART00000109390
|
AL954361.1
|
|
| chr1_-_35653560 | 9.40 |
ENSDART00000142154
|
frem3
|
Fras1 related extracellular matrix 3 |
| chr7_-_22956889 | 9.14 |
ENSDART00000101447
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr13_+_28821841 | 9.01 |
ENSDART00000179900
|
CU639469.1
|
|
| chr9_-_43073960 | 8.96 |
ENSDART00000059460
|
ttn.2
|
titin, tandem duplicate 2 |
| chr20_+_6756247 | 8.62 |
ENSDART00000167344
|
igfbp3
|
insulin-like growth factor binding protein 3 |
| chr14_+_30328567 | 8.27 |
ENSDART00000105889
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr12_-_19119176 | 8.26 |
ENSDART00000149180
|
aco2
|
aconitase 2, mitochondrial |
| chr8_-_32497815 | 8.06 |
ENSDART00000122359
|
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr16_-_31475904 | 8.04 |
ENSDART00000145691
|
si:ch211-251p5.5
|
si:ch211-251p5.5 |
| chr3_-_50046004 | 7.86 |
ENSDART00000109544
|
si:ch1073-100f3.2
|
si:ch1073-100f3.2 |
| chr23_+_32039386 | 7.84 |
ENSDART00000133801
|
mylk2
|
myosin light chain kinase 2 |
| chr14_-_8795081 | 7.81 |
ENSDART00000106671
|
tgfb2l
|
transforming growth factor, beta 2, like |
| chr13_+_38817871 | 7.80 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr21_+_40589770 | 7.73 |
ENSDART00000164650
ENSDART00000161584 ENSDART00000161108 |
pdk3b
|
pyruvate dehydrogenase kinase, isozyme 3b |
| chr23_+_36601984 | 7.57 |
ENSDART00000128598
|
igfbp6b
|
insulin-like growth factor binding protein 6b |
| chr3_-_27915270 | 7.32 |
ENSDART00000115370
|
mettl22
|
methyltransferase like 22 |
| chr16_+_34046733 | 7.26 |
ENSDART00000058424
|
fam46ba
|
family with sequence similarity 46, member Ba |
| chr5_+_69808763 | 7.25 |
ENSDART00000143482
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr13_-_30149973 | 7.20 |
ENSDART00000041515
|
sar1ab
|
secretion associated, Ras related GTPase 1Ab |
| chrM_-_15232 | 7.16 |
ENSDART00000093623
|
mt-nd6
|
NADH dehydrogenase 6, mitochondrial |
| chr23_-_39849155 | 6.95 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr16_-_29164379 | 6.93 |
ENSDART00000132589
|
mef2d
|
myocyte enhancer factor 2d |
| chr14_-_46451249 | 6.89 |
ENSDART00000058466
|
fgfbp2a
|
fibroblast growth factor binding protein 2a |
| chr1_-_13271085 | 6.46 |
ENSDART00000193663
|
pcdh18a
|
protocadherin 18a |
| chr6_-_46768040 | 6.19 |
ENSDART00000154071
|
igfn1.2
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 2 |
| chr5_-_8711157 | 6.15 |
ENSDART00000062957
|
paip1
|
poly(A) binding protein interacting protein 1 |
| chr22_-_7006974 | 5.99 |
ENSDART00000133143
|
gpd1b
|
glycerol-3-phosphate dehydrogenase 1b |
| chr8_-_32497581 | 5.91 |
ENSDART00000176298
ENSDART00000183340 |
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr3_+_28939759 | 5.87 |
ENSDART00000141904
|
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr10_-_17232372 | 5.86 |
ENSDART00000135679
|
rab36
|
RAB36, member RAS oncogene family |
| chr20_+_33532296 | 5.83 |
ENSDART00000153153
|
kcnf1a
|
potassium voltage-gated channel, subfamily F, member 1a |
| chr19_-_12648122 | 5.81 |
ENSDART00000151184
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr15_+_28940352 | 5.33 |
ENSDART00000154085
|
gipr
|
gastric inhibitory polypeptide receptor |
| chr1_-_13271569 | 5.24 |
ENSDART00000127838
|
pcdh18a
|
protocadherin 18a |
| chr6_-_10863582 | 5.21 |
ENSDART00000135454
ENSDART00000133893 ENSDART00000014521 ENSDART00000164342 |
lnpk
|
lunapark, ER junction formation factor |
| chr5_-_32505109 | 5.15 |
ENSDART00000188219
|
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr9_-_23990416 | 5.10 |
ENSDART00000113176
|
col6a3
|
collagen, type VI, alpha 3 |
| chr16_-_17541890 | 5.09 |
ENSDART00000131328
|
clcn1b
|
chloride channel, voltage-sensitive 1b |
| chr18_-_16791331 | 4.94 |
ENSDART00000148222
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr7_-_22956716 | 4.93 |
ENSDART00000122113
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr8_-_18010735 | 4.78 |
ENSDART00000125014
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr18_-_11184584 | 4.77 |
ENSDART00000040500
|
tspan9a
|
tetraspanin 9a |
| chr14_+_23518110 | 4.67 |
ENSDART00000112930
|
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr10_-_10607118 | 4.64 |
ENSDART00000101089
|
dbh
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr10_+_36345176 | 4.61 |
ENSDART00000099397
|
or105-1
|
odorant receptor, family C, subfamily 105, member 1 |
| chr1_-_22652424 | 4.59 |
ENSDART00000036797
|
uchl1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr12_+_27537357 | 4.57 |
ENSDART00000136212
|
etv4
|
ets variant 4 |
| chr12_+_18718065 | 4.45 |
ENSDART00000152935
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr23_+_14097508 | 4.43 |
ENSDART00000087280
|
cacnb3a
|
calcium channel, voltage-dependent, beta 3a |
| chr5_-_32505276 | 4.36 |
ENSDART00000034705
ENSDART00000187597 |
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr7_+_39006837 | 4.29 |
ENSDART00000173735
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr6_-_52234796 | 4.28 |
ENSDART00000001803
|
tomm34
|
translocase of outer mitochondrial membrane 34 |
| chr17_+_5976683 | 4.27 |
ENSDART00000110276
|
zgc:194275
|
zgc:194275 |
| chr22_-_8509215 | 4.27 |
ENSDART00000140146
|
si:ch73-27e22.3
|
si:ch73-27e22.3 |
| chr14_+_23520986 | 4.25 |
ENSDART00000170473
ENSDART00000175970 |
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr7_-_44957503 | 4.24 |
ENSDART00000165077
|
cdh16
|
cadherin 16, KSP-cadherin |
| chr9_+_27876146 | 4.17 |
ENSDART00000133997
|
armc8
|
armadillo repeat containing 8 |
| chr2_-_28102264 | 3.94 |
ENSDART00000013638
|
cdh6
|
cadherin 6 |
| chr7_+_40093662 | 3.82 |
ENSDART00000053011
|
si:ch73-174h16.4
|
si:ch73-174h16.4 |
| chr16_-_32837806 | 3.79 |
ENSDART00000003997
|
MCHR2
|
si:dkey-165n16.5 |
| chr11_+_41560792 | 3.77 |
ENSDART00000127292
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr1_+_33969015 | 3.69 |
ENSDART00000042984
ENSDART00000146530 |
epha6
|
eph receptor A6 |
| chr16_-_32975951 | 3.64 |
ENSDART00000101969
ENSDART00000175149 |
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr8_+_21159122 | 3.58 |
ENSDART00000033491
|
spryd4
|
SPRY domain containing 4 |
| chr7_+_57677120 | 3.55 |
ENSDART00000110623
|
arsj
|
arylsulfatase family, member J |
| chr17_-_5769196 | 3.52 |
ENSDART00000113885
|
si:dkey-100n19.2
|
si:dkey-100n19.2 |
| chr22_-_13649588 | 3.50 |
ENSDART00000131877
|
si:ch211-279g13.1
|
si:ch211-279g13.1 |
| chr6_-_49873020 | 3.45 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr18_+_10820430 | 3.43 |
ENSDART00000161990
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr15_+_15856178 | 3.40 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr21_-_39253647 | 3.38 |
ENSDART00000133034
|
tusc5b
|
tumor suppressor candidate 5b |
| chr1_-_8917902 | 3.32 |
ENSDART00000137900
|
grin2ab
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A, b |
| chr25_-_6557854 | 3.21 |
ENSDART00000181740
|
cspg4
|
chondroitin sulfate proteoglycan 4 |
| chr18_-_38087875 | 3.21 |
ENSDART00000111301
|
luzp2
|
leucine zipper protein 2 |
| chr25_+_19105804 | 3.13 |
ENSDART00000104414
|
rlbp1b
|
retinaldehyde binding protein 1b |
| chr10_-_17501528 | 3.03 |
ENSDART00000144847
|
slc2a11l
|
solute carrier family 2 (facilitated glucose transporter), member 11-like |
| chr25_-_15245475 | 3.02 |
ENSDART00000142943
|
ccdc73
|
coiled-coil domain containing 73 |
| chr16_-_16590489 | 2.92 |
ENSDART00000190021
|
si:ch211-257p13.3
|
si:ch211-257p13.3 |
| chr16_-_36798783 | 2.88 |
ENSDART00000145697
|
calb1
|
calbindin 1 |
| chr2_-_5356686 | 2.85 |
ENSDART00000124290
|
MFN1
|
mitofusin 1 |
| chr5_-_64511428 | 2.83 |
ENSDART00000016321
|
rxrab
|
retinoid x receptor, alpha b |
| chr12_+_27141140 | 2.80 |
ENSDART00000136415
|
hoxb1b
|
homeobox B1b |
| chr24_-_27409599 | 2.77 |
ENSDART00000041770
|
ccl34b.3
|
chemokine (C-C motif) ligand 34b, duplicate 3 |
| chr20_+_2134816 | 2.75 |
ENSDART00000039249
|
l3mbtl3
|
l(3)mbt-like 3 (Drosophila) |
| chr17_+_8184649 | 2.64 |
ENSDART00000091818
|
tulp4b
|
tubby like protein 4b |
| chr24_-_5691956 | 2.64 |
ENSDART00000189112
|
dia1b
|
deleted in autism 1b |
| chr1_-_45662774 | 2.61 |
ENSDART00000042158
|
serhl
|
serine hydrolase-like |
| chr17_-_16069905 | 2.54 |
ENSDART00000110383
|
map7a
|
microtubule-associated protein 7a |
| chr18_+_8339596 | 2.54 |
ENSDART00000183323
ENSDART00000185397 |
BX276107.1
|
|
| chr12_-_6905375 | 2.51 |
ENSDART00000152322
|
pcdh15b
|
protocadherin-related 15b |
| chr12_-_20340543 | 2.50 |
ENSDART00000055623
|
hbbe3
|
hemoglobin beta embryonic-3 |
| chr24_-_6647275 | 2.44 |
ENSDART00000161494
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr20_-_2134620 | 2.43 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr10_+_41765616 | 2.42 |
ENSDART00000170682
|
rnf34b
|
ring finger protein 34b |
| chr25_+_3677650 | 2.34 |
ENSDART00000154348
|
prnprs3
|
prion protein, related sequence 3 |
| chr2_-_36925561 | 2.28 |
ENSDART00000187690
|
map1sb
|
microtubule-associated protein 1Sb |
| chr20_+_812012 | 2.24 |
ENSDART00000179299
ENSDART00000137135 |
mei4
|
meiosis-specific, MEI4 homolog (S. cerevisiae) |
| chr2_-_3678029 | 2.17 |
ENSDART00000146861
|
mmp16b
|
matrix metallopeptidase 16b (membrane-inserted) |
| chr4_-_2719128 | 2.15 |
ENSDART00000184693
|
slco1c1
|
solute carrier organic anion transporter family, member 1C1 |
| chr23_-_19953089 | 2.14 |
ENSDART00000153828
|
atp2b3b
|
ATPase plasma membrane Ca2+ transporting 3b |
| chr12_+_29236274 | 2.13 |
ENSDART00000006505
|
mxtx2
|
mix-type homeobox gene 2 |
| chr1_+_20084389 | 2.06 |
ENSDART00000140263
|
prss12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr13_-_33114933 | 2.06 |
ENSDART00000140543
ENSDART00000075953 |
zfyve26
|
zinc finger, FYVE domain containing 26 |
| chr23_-_7797207 | 2.03 |
ENSDART00000181611
|
myt1b
|
myelin transcription factor 1b |
| chr17_-_23221504 | 2.02 |
ENSDART00000156411
|
fam98a
|
family with sequence similarity 98, member A |
| chr19_+_9091673 | 1.98 |
ENSDART00000052898
|
si:ch211-81a5.5
|
si:ch211-81a5.5 |
| chr1_-_42778510 | 1.98 |
ENSDART00000190172
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr5_+_24305877 | 1.96 |
ENSDART00000144226
|
ctsll
|
cathepsin L, like |
| chr2_+_35880600 | 1.95 |
ENSDART00000004277
|
lamc1
|
laminin, gamma 1 |
| chr9_-_22188117 | 1.91 |
ENSDART00000132890
|
crygm2d17
|
crystallin, gamma M2d17 |
| chr6_-_18698456 | 1.86 |
ENSDART00000166587
|
rhbdl3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr20_+_6630540 | 1.85 |
ENSDART00000138361
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr23_-_27633730 | 1.84 |
ENSDART00000103639
|
arf3a
|
ADP-ribosylation factor 3a |
| chr17_-_34853229 | 1.83 |
ENSDART00000152033
|
kidins220a
|
kinase D-interacting substrate 220a |
| chr21_-_22910520 | 1.81 |
ENSDART00000065567
ENSDART00000191792 |
guca1d
|
guanylate cyclase activator 1d |
| chr6_-_894006 | 1.80 |
ENSDART00000171091
|
zeb2b
|
zinc finger E-box binding homeobox 2b |
| chr15_-_34668485 | 1.74 |
ENSDART00000186605
|
bag6
|
BCL2 associated athanogene 6 |
| chr4_+_9279515 | 1.69 |
ENSDART00000048707
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr3_-_34100700 | 1.65 |
ENSDART00000151628
|
ighv6-1
|
immunoglobulin heavy variable 6-1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mef2aa+mef2ab
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.4 | 28.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 8.8 | 35.1 | GO:0001778 | plasma membrane repair(GO:0001778) chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 8.6 | 42.8 | GO:0090200 | regulation of mitochondrial membrane potential(GO:0051881) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 8.2 | 24.6 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 6.5 | 148.9 | GO:0031033 | myosin filament organization(GO:0031033) |
| 4.8 | 24.2 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 4.4 | 39.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 3.8 | 11.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 3.7 | 52.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 3.1 | 40.3 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 2.9 | 14.7 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 2.8 | 119.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 2.7 | 10.7 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 2.3 | 49.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 1.8 | 7.2 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 1.8 | 14.1 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 1.7 | 5.2 | GO:1903373 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 1.7 | 25.6 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 1.6 | 7.9 | GO:0015743 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 1.5 | 4.6 | GO:0042421 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 1.5 | 6.0 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 1.5 | 16.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 1.4 | 12.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 1.1 | 19.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 1.1 | 12.6 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.9 | 12.1 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.8 | 1.6 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.7 | 2.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.7 | 23.7 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.7 | 2.9 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.7 | 16.9 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.6 | 11.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.6 | 12.0 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.6 | 10.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.6 | 18.2 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.5 | 2.0 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.4 | 16.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.4 | 19.9 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.4 | 2.8 | GO:0021570 | rhombomere 4 development(GO:0021570) rhombomere 4 morphogenesis(GO:0021661) |
| 0.4 | 3.8 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.4 | 4.5 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.4 | 3.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.4 | 23.9 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.3 | 2.4 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.3 | 2.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.3 | 11.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.3 | 4.6 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.3 | 163.1 | GO:0061061 | muscle structure development(GO:0061061) |
| 0.3 | 1.0 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.3 | 8.3 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.3 | 2.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.3 | 1.1 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 0.2 | 23.1 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.2 | 6.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.2 | 29.4 | GO:0006821 | chloride transport(GO:0006821) |
| 0.2 | 7.7 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.2 | 4.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 3.3 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.2 | 3.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 11.2 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.1 | 1.2 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.1 | 2.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 4.1 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 4.3 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.1 | 1.8 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 7.7 | GO:0071559 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.1 | 2.3 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 6.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 10.8 | GO:0021782 | glial cell development(GO:0021782) |
| 0.1 | 0.4 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 7.3 | GO:0008213 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.1 | 1.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 53.4 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.1 | 1.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 2.8 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 1.4 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 3.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 3.3 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.1 | 1.4 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.1 | 1.5 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 4.1 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 3.4 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 2.1 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 4.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 2.8 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.3 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.9 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.6 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 4.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 2.1 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 5.9 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 3.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.2 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 7.1 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.7 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 1.9 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.9 | GO:0035107 | appendage morphogenesis(GO:0035107) |
| 0.0 | 1.6 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.2 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.1 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 96.8 | GO:0031430 | M band(GO:0031430) |
| 3.1 | 134.0 | GO:0005861 | troponin complex(GO:0005861) |
| 2.7 | 127.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 2.6 | 39.3 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 2.1 | 24.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 2.0 | 19.9 | GO:0031672 | A band(GO:0031672) |
| 1.7 | 5.2 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.9 | 35.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.8 | 4.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.7 | 4.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.7 | 27.3 | GO:0016605 | PML body(GO:0016605) |
| 0.6 | 19.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.5 | 12.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.4 | 10.7 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.4 | 29.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.3 | 2.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 3.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.3 | 2.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 1.7 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.2 | 3.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 4.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 17.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.2 | 2.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 4.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 9.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 14.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 19.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 7.4 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 2.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 1.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 9.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 3.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 60.8 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.1 | 4.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 5.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 22.7 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.1 | 1.1 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 1.3 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
| 0.0 | 1.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 2.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 8.5 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 8.7 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 9.7 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 76.7 | GO:0005634 | nucleus(GO:0005634) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.2 | 24.6 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 6.8 | 27.3 | GO:0005521 | lamin binding(GO:0005521) |
| 6.4 | 127.4 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 4.7 | 52.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 4.6 | 18.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 3.1 | 40.3 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 3.0 | 45.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 2.8 | 8.3 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 2.5 | 54.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.8 | 18.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 1.6 | 7.9 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 1.5 | 4.6 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 1.5 | 6.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 1.5 | 16.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 1.4 | 11.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 1.3 | 10.7 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 1.3 | 7.8 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 1.1 | 11.5 | GO:0005522 | profilin binding(GO:0005522) |
| 1.1 | 12.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 1.1 | 5.3 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 1.0 | 7.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.9 | 26.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.9 | 16.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.9 | 28.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.9 | 3.6 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.9 | 24.9 | GO:0016208 | AMP binding(GO:0016208) |
| 0.9 | 2.6 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.8 | 17.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.8 | 7.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.8 | 52.6 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.7 | 35.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.7 | 34.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.7 | 3.5 | GO:0050780 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.7 | 6.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.7 | 4.8 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.7 | 10.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.6 | 7.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.5 | 42.7 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.5 | 1.4 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.4 | 13.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.3 | 9.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.3 | 2.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 21.8 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.3 | 29.4 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.3 | 2.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.3 | 4.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 229.4 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.2 | 4.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.2 | 3.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 3.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.2 | 2.5 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.2 | 3.8 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.2 | 3.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 3.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 21.5 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.2 | 4.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 4.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 6.4 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.1 | 1.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.4 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.6 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 4.3 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 10.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.6 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 23.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 7.4 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.1 | 10.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 3.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 3.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 2.8 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 4.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.3 | GO:1990939 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.1 | 18.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.7 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 5.8 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 2.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.0 | 10.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 2.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.3 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 47.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.6 | 22.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 7.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 15.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.3 | 11.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.3 | 6.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 16.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 4.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 8.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 3.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 1.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 1.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 2.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 47.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 1.1 | 8.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 1.0 | 19.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 1.0 | 28.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.0 | 11.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.9 | 18.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.7 | 10.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.5 | 22.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.4 | 4.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.4 | 6.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.4 | 8.8 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.3 | 3.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.3 | 2.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.3 | 3.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 3.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 6.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.7 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 3.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.7 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |