Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
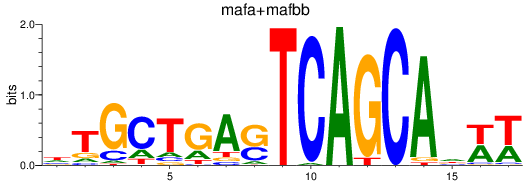
Results for mafa+mafbb
Z-value: 1.39
Transcription factors associated with mafa+mafbb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mafa
|
ENSDARG00000015890 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
|
mafbb
|
ENSDARG00000070542 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Bb |
|
mafbb
|
ENSDARG00000099120 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Bb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mafbb | dr11_v1_chr11_-_25384213_25384213 | 0.20 | 5.4e-02 | Click! |
| mafa | dr11_v1_chr18_+_29403017_29403017 | 0.18 | 7.5e-02 | Click! |
Activity profile of mafa+mafbb motif
Sorted Z-values of mafa+mafbb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_18030938 | 33.90 |
ENSDART00000013540
|
si:ch73-141c7.1
|
si:ch73-141c7.1 |
| chr13_-_20381485 | 31.01 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr22_-_24738188 | 23.83 |
ENSDART00000050238
|
vtg1
|
vitellogenin 1 |
| chr2_+_25816843 | 21.26 |
ENSDART00000078639
|
slc2a2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr10_+_10801564 | 20.16 |
ENSDART00000027026
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr10_+_10801719 | 19.59 |
ENSDART00000193648
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr24_-_12938922 | 19.40 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr13_+_2908764 | 19.33 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr22_-_24880824 | 18.22 |
ENSDART00000061165
|
vtg2
|
vitellogenin 2 |
| chr22_-_24757785 | 17.53 |
ENSDART00000078225
|
vtg5
|
vitellogenin 5 |
| chr10_+_28428222 | 16.65 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr15_+_21202820 | 13.90 |
ENSDART00000154036
|
si:dkey-52d15.2
|
si:dkey-52d15.2 |
| chr23_-_31428763 | 12.75 |
ENSDART00000053545
|
zgc:153284
|
zgc:153284 |
| chr10_+_38708099 | 11.77 |
ENSDART00000172306
|
tmprss2
|
transmembrane protease, serine 2 |
| chr20_+_38285671 | 10.90 |
ENSDART00000061432
|
ccl38a.4
|
chemokine (C-C motif) ligand 38, duplicate 4 |
| chr16_+_18974064 | 10.44 |
ENSDART00000079248
|
slc6a19b
|
solute carrier family 6 (neutral amino acid transporter), member 19b |
| chr11_-_5865744 | 10.19 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr13_-_25196758 | 10.17 |
ENSDART00000184722
|
adka
|
adenosine kinase a |
| chr5_-_37116265 | 10.00 |
ENSDART00000057613
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr10_+_26747755 | 9.92 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr19_-_35428815 | 9.67 |
ENSDART00000169006
ENSDART00000003167 |
ak2
|
adenylate kinase 2 |
| chr5_-_30615901 | 9.26 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr1_-_54706039 | 8.38 |
ENSDART00000083633
|
exosc1
|
exosome component 1 |
| chr16_+_46294337 | 8.36 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr16_-_12319822 | 8.30 |
ENSDART00000127453
ENSDART00000184526 |
trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr12_-_44043285 | 8.07 |
ENSDART00000163074
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr17_-_53359028 | 7.97 |
ENSDART00000185218
|
CABZ01068208.1
|
|
| chr19_+_7567763 | 7.52 |
ENSDART00000140411
|
s100a11
|
S100 calcium binding protein A11 |
| chr21_-_43328056 | 7.44 |
ENSDART00000114955
|
sowahaa
|
sosondowah ankyrin repeat domain family member Aa |
| chr11_-_40128722 | 7.29 |
ENSDART00000165781
|
fam83e
|
family with sequence similarity 83, member E |
| chr11_-_33960318 | 7.10 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr12_-_3940768 | 7.06 |
ENSDART00000134292
|
zgc:92040
|
zgc:92040 |
| chr10_+_10387328 | 6.61 |
ENSDART00000080904
|
sardh
|
sarcosine dehydrogenase |
| chr8_-_554540 | 6.56 |
ENSDART00000163934
|
FO704758.1
|
Danio rerio uncharacterized LOC100329294 (LOC100329294), mRNA. |
| chr16_+_46410520 | 6.38 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr11_-_23501467 | 6.20 |
ENSDART00000169066
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr17_-_20236228 | 6.19 |
ENSDART00000136490
ENSDART00000029380 |
bnip4
|
BCL2 interacting protein 4 |
| chr24_-_42090635 | 6.15 |
ENSDART00000166413
|
ssr1
|
signal sequence receptor, alpha |
| chr19_+_1465004 | 6.08 |
ENSDART00000159157
|
CABZ01073736.1
|
|
| chr12_+_34258139 | 6.04 |
ENSDART00000153127
|
socs3b
|
suppressor of cytokine signaling 3b |
| chr4_+_68562464 | 6.02 |
ENSDART00000192954
|
BX548011.4
|
|
| chr13_-_31025505 | 5.92 |
ENSDART00000137709
|
wdfy4
|
WDFY family member 4 |
| chr21_-_28523548 | 5.87 |
ENSDART00000077910
|
epdl2
|
ependymin-like 2 |
| chr21_+_43199237 | 5.72 |
ENSDART00000151748
|
aff4
|
AF4/FMR2 family, member 4 |
| chr6_-_26895750 | 5.63 |
ENSDART00000011863
|
hdlbpa
|
high density lipoprotein binding protein a |
| chr8_-_41228530 | 5.51 |
ENSDART00000165949
ENSDART00000173055 |
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr5_+_9377005 | 5.48 |
ENSDART00000124924
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr17_-_53353653 | 5.46 |
ENSDART00000180744
ENSDART00000026879 |
unm_sa911
|
un-named sa911 |
| chr23_+_7518294 | 5.31 |
ENSDART00000081536
|
hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr20_-_43079478 | 5.27 |
ENSDART00000060982
|
snx9b
|
sorting nexin 9b |
| chr14_+_3507326 | 5.23 |
ENSDART00000159326
|
gstp1
|
glutathione S-transferase pi 1 |
| chr4_-_71708855 | 5.23 |
ENSDART00000158819
|
si:dkeyp-4f2.1
|
si:dkeyp-4f2.1 |
| chr19_+_48024457 | 5.09 |
ENSDART00000163823
|
kpnb1
|
karyopherin (importin) beta 1 |
| chr17_+_6217704 | 5.03 |
ENSDART00000129100
|
BX000363.1
|
|
| chr1_-_52498146 | 5.00 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr18_+_10926197 | 4.92 |
ENSDART00000192387
|
ttc38
|
tetratricopeptide repeat domain 38 |
| chr12_+_20552190 | 4.85 |
ENSDART00000113224
|
gna13a
|
guanine nucleotide binding protein (G protein), alpha 13a |
| chr5_-_65004578 | 4.74 |
ENSDART00000169287
|
ANXA1 (1 of many)
|
zgc:110283 |
| chr24_-_26369185 | 4.72 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr13_-_25199260 | 4.66 |
ENSDART00000057605
|
adka
|
adenosine kinase a |
| chr14_+_146857 | 4.61 |
ENSDART00000122521
|
CABZ01088229.1
|
|
| chr9_-_55586151 | 4.53 |
ENSDART00000181886
|
arsh
|
arylsulfatase H |
| chr9_+_28116079 | 4.49 |
ENSDART00000101338
|
idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr14_-_49859747 | 4.37 |
ENSDART00000169456
ENSDART00000164967 |
nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr24_+_19518303 | 4.36 |
ENSDART00000027022
ENSDART00000056080 |
sulf1
|
sulfatase 1 |
| chr8_-_22538588 | 4.33 |
ENSDART00000144041
|
csde1
|
cold shock domain containing E1, RNA-binding |
| chr20_-_14054083 | 4.32 |
ENSDART00000009549
|
rhag
|
Rh associated glycoprotein |
| chr23_-_16666583 | 4.21 |
ENSDART00000189804
ENSDART00000136496 |
sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr5_-_65004791 | 4.21 |
ENSDART00000180023
|
ANXA1 (1 of many)
|
zgc:110283 |
| chr5_-_32338866 | 4.14 |
ENSDART00000017956
ENSDART00000047670 |
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr1_-_52497834 | 3.97 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr7_+_61764040 | 3.88 |
ENSDART00000056745
|
acox3
|
acyl-CoA oxidase 3, pristanoyl |
| chr20_+_46385907 | 3.82 |
ENSDART00000060710
|
adgrg11
|
adhesion G protein-coupled receptor G11 |
| chr8_+_31435452 | 3.79 |
ENSDART00000145282
|
selenop
|
selenoprotein P |
| chr24_-_23998897 | 3.75 |
ENSDART00000130053
|
zmp:0000000991
|
zmp:0000000991 |
| chr4_-_71708567 | 3.72 |
ENSDART00000182645
|
si:dkeyp-4f2.1
|
si:dkeyp-4f2.1 |
| chr3_+_23063718 | 3.71 |
ENSDART00000140225
ENSDART00000184431 |
b4galnt2.1
|
beta-1,4-N-acetyl-galactosaminyl transferase 2, tandem duplicate 1 |
| chr7_-_19043401 | 3.62 |
ENSDART00000158796
|
fcer1g
|
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide |
| chr7_+_3442834 | 3.60 |
ENSDART00000138247
|
si:ch211-285c6.2
|
si:ch211-285c6.2 |
| chr18_-_45617146 | 3.60 |
ENSDART00000146543
|
wt1b
|
wilms tumor 1b |
| chr2_+_19163965 | 3.58 |
ENSDART00000166073
|
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr3_+_14463941 | 3.49 |
ENSDART00000170927
|
cnn1b
|
calponin 1, basic, smooth muscle, b |
| chr1_-_30457062 | 3.43 |
ENSDART00000185318
ENSDART00000157924 ENSDART00000161380 |
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr1_-_30510839 | 3.42 |
ENSDART00000168189
ENSDART00000174868 |
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr5_+_38684651 | 3.40 |
ENSDART00000137852
|
si:dkey-58f10.10
|
si:dkey-58f10.10 |
| chr8_+_28629741 | 3.34 |
ENSDART00000163915
|
si:dkey-109a10.2
|
si:dkey-109a10.2 |
| chr15_+_34069746 | 3.34 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr23_+_42304602 | 3.34 |
ENSDART00000166004
|
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr8_+_50190742 | 3.31 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr5_+_38679623 | 3.27 |
ENSDART00000131879
ENSDART00000170010 |
si:dkey-58f10.11
si:dkey-58f10.10
|
si:dkey-58f10.11 si:dkey-58f10.10 |
| chr23_-_44848961 | 3.27 |
ENSDART00000136839
|
wu:fb72h05
|
wu:fb72h05 |
| chr3_-_7948799 | 3.26 |
ENSDART00000163714
|
trim35-24
|
tripartite motif containing 35-24 |
| chr10_-_29120515 | 3.15 |
ENSDART00000162016
ENSDART00000149140 |
zpld1a
|
zona pellucida-like domain containing 1a |
| chr1_+_54766943 | 3.14 |
ENSDART00000144759
|
nlrc6
|
NLR family CARD domain containing 6 |
| chr5_-_67471375 | 3.08 |
ENSDART00000147009
|
si:dkey-251i10.2
|
si:dkey-251i10.2 |
| chr22_-_263117 | 3.05 |
ENSDART00000158134
|
zgc:66156
|
zgc:66156 |
| chr16_-_51299061 | 3.03 |
ENSDART00000148677
|
serpinb1l4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 4 |
| chr21_+_19347655 | 3.02 |
ENSDART00000093155
|
hpse
|
heparanase |
| chr18_+_14595805 | 2.99 |
ENSDART00000167825
|
wfdc1
|
WAP four-disulfide core domain 1 |
| chr7_-_25697285 | 2.99 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr15_+_41932069 | 2.99 |
ENSDART00000143007
|
si:ch211-191a16.2
|
si:ch211-191a16.2 |
| chr16_-_16481046 | 2.95 |
ENSDART00000158704
|
nbeal2
|
neurobeachin-like 2 |
| chr15_+_3790556 | 2.94 |
ENSDART00000181467
|
RNF14 (1 of many)
|
ring finger protein 14 |
| chr16_+_23326085 | 2.94 |
ENSDART00000180935
|
krtcap2
|
keratinocyte associated protein 2 |
| chr7_+_57108823 | 2.93 |
ENSDART00000184943
ENSDART00000055956 |
enosf1
|
enolase superfamily member 1 |
| chr14_-_25928541 | 2.91 |
ENSDART00000145850
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr20_-_23852174 | 2.82 |
ENSDART00000122414
|
si:dkey-15j16.6
|
si:dkey-15j16.6 |
| chr15_-_41762530 | 2.82 |
ENSDART00000187125
ENSDART00000154971 |
ftr91
|
finTRIM family, member 91 |
| chr10_+_8527196 | 2.80 |
ENSDART00000141147
|
si:ch211-193e13.5
|
si:ch211-193e13.5 |
| chr4_-_58964138 | 2.79 |
ENSDART00000150259
|
si:ch211-64i20.3
|
si:ch211-64i20.3 |
| chr16_-_33806390 | 2.73 |
ENSDART00000160671
|
rspo1
|
R-spondin 1 |
| chr4_-_70131926 | 2.68 |
ENSDART00000108511
|
si:ch211-208f21.3
|
si:ch211-208f21.3 |
| chr4_+_77709678 | 2.66 |
ENSDART00000036856
|
AL935186.1
|
|
| chr12_+_16967715 | 2.65 |
ENSDART00000138174
|
slc16a12b
|
solute carrier family 16, member 12b |
| chr7_+_27317174 | 2.59 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr2_-_38256589 | 2.57 |
ENSDART00000173064
|
si:ch211-14a17.10
|
si:ch211-14a17.10 |
| chr6_-_35439406 | 2.42 |
ENSDART00000073784
|
rgs5a
|
regulator of G protein signaling 5a |
| chr8_+_23439340 | 2.36 |
ENSDART00000109932
ENSDART00000185469 |
foxp3b
|
forkhead box P3b |
| chr22_-_24285432 | 2.35 |
ENSDART00000164083
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr6_-_426041 | 2.33 |
ENSDART00000162789
|
fam83fb
|
family with sequence similarity 83, member Fb |
| chr19_-_7441686 | 2.30 |
ENSDART00000168194
|
gabpb2a
|
GA binding protein transcription factor, beta subunit 2a |
| chr7_+_35068036 | 2.30 |
ENSDART00000022139
|
zgc:136461
|
zgc:136461 |
| chr12_-_44019561 | 2.29 |
ENSDART00000165900
|
si:dkey-31i7.1
|
si:dkey-31i7.1 |
| chr5_-_26893310 | 2.28 |
ENSDART00000126669
|
lman2lb
|
lectin, mannose-binding 2-like b |
| chr19_+_23308419 | 2.24 |
ENSDART00000141558
|
irgf2
|
immunity-related GTPase family, f2 |
| chr18_+_22302635 | 2.17 |
ENSDART00000141051
|
carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr7_-_6415991 | 2.15 |
ENSDART00000173349
|
CU457819.3
|
Histone H3.2 |
| chr4_+_5317483 | 2.12 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr14_-_4076480 | 2.11 |
ENSDART00000059231
|
enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr4_+_65057234 | 2.09 |
ENSDART00000162985
|
si:dkey-14o6.2
|
si:dkey-14o6.2 |
| chr11_+_42641404 | 2.06 |
ENSDART00000172641
ENSDART00000169938 |
il17rd
|
interleukin 17 receptor D |
| chr4_-_67909331 | 2.04 |
ENSDART00000191908
|
si:ch211-66c13.1
|
si:ch211-66c13.1 |
| chr14_-_7786585 | 2.04 |
ENSDART00000186440
|
tmem173
|
transmembrane protein 173 |
| chr20_-_20410029 | 2.02 |
ENSDART00000192177
ENSDART00000063483 |
prkchb
|
protein kinase C, eta, b |
| chr4_+_42878556 | 2.02 |
ENSDART00000171047
|
si:zfos-451d2.2
|
si:zfos-451d2.2 |
| chr18_-_6151793 | 2.02 |
ENSDART00000122307
|
gcsha
|
glycine cleavage system protein H (aminomethyl carrier), a |
| chr11_+_42422638 | 2.01 |
ENSDART00000042599
ENSDART00000181175 |
dennd6aa
|
DENN/MADD domain containing 6Aa |
| chr9_+_32930622 | 2.01 |
ENSDART00000100928
|
cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr19_-_43639331 | 1.96 |
ENSDART00000138009
ENSDART00000086138 |
fam83hb
|
family with sequence similarity 83, member Hb |
| chr3_-_33997763 | 1.94 |
ENSDART00000151461
ENSDART00000151307 |
ighz
|
immunoglobulin heavy constant zeta |
| chr16_+_17089114 | 1.90 |
ENSDART00000173223
|
cd27
|
CD27 molecule |
| chr5_+_18014931 | 1.90 |
ENSDART00000142562
|
ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr3_-_19517462 | 1.82 |
ENSDART00000162027
|
CU571315.2
|
|
| chr1_+_29862074 | 1.82 |
ENSDART00000133905
ENSDART00000136786 ENSDART00000135252 |
pcca
|
propionyl CoA carboxylase, alpha polypeptide |
| chr5_+_38685089 | 1.79 |
ENSDART00000139743
|
si:dkey-58f10.10
|
si:dkey-58f10.10 |
| chr10_-_40462894 | 1.77 |
ENSDART00000147924
|
taar20b1
|
trace amine associated receptor 20b1 |
| chr25_-_13558172 | 1.73 |
ENSDART00000139970
ENSDART00000133789 ENSDART00000144426 |
ano10b
|
anoctamin 10b |
| chr20_-_39219537 | 1.71 |
ENSDART00000005764
|
cyp39a1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr17_-_19345521 | 1.65 |
ENSDART00000082085
|
gsc
|
goosecoid |
| chr4_+_47257854 | 1.63 |
ENSDART00000173868
|
crestin
|
crestin |
| chr1_+_19303241 | 1.60 |
ENSDART00000129970
|
si:dkeyp-118a3.2
|
si:dkeyp-118a3.2 |
| chr20_-_16972351 | 1.59 |
ENSDART00000148312
ENSDART00000186702 |
si:ch73-74h11.1
|
si:ch73-74h11.1 |
| chr8_+_23436411 | 1.57 |
ENSDART00000140044
|
foxp3b
|
forkhead box P3b |
| chr23_+_24124684 | 1.57 |
ENSDART00000144478
|
si:dkey-21o19.2
|
si:dkey-21o19.2 |
| chr23_+_30707837 | 1.54 |
ENSDART00000016096
|
dnajc11a
|
DnaJ (Hsp40) homolog, subfamily C, member 11a |
| chr9_+_13985567 | 1.52 |
ENSDART00000102296
|
cd28
|
CD28 molecule |
| chr5_-_43959972 | 1.50 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr2_-_4496358 | 1.47 |
ENSDART00000158216
|
adipoqb
|
adiponectin, C1Q and collagen domain containing, b |
| chr23_+_7379728 | 1.46 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr9_+_6802641 | 1.46 |
ENSDART00000187278
|
FO203514.1
|
|
| chr4_-_36032930 | 1.45 |
ENSDART00000191414
|
zgc:174180
|
zgc:174180 |
| chr14_-_24001825 | 1.44 |
ENSDART00000130704
|
il4
|
interleukin 4 |
| chr4_-_42397126 | 1.43 |
ENSDART00000162437
|
si:ch211-129p6.2
|
si:ch211-129p6.2 |
| chr4_+_34121902 | 1.41 |
ENSDART00000170225
|
si:ch211-223g7.6
|
si:ch211-223g7.6 |
| chr19_-_7441948 | 1.40 |
ENSDART00000003544
|
gabpb2a
|
GA binding protein transcription factor, beta subunit 2a |
| chr10_-_40514643 | 1.40 |
ENSDART00000140705
|
taar19k
|
trace amine associated receptor 19k |
| chr6_-_13783604 | 1.38 |
ENSDART00000149536
ENSDART00000041269 ENSDART00000150102 |
cryba2a
|
crystallin, beta A2a |
| chr6_-_29515709 | 1.34 |
ENSDART00000180205
|
pex5la
|
peroxisomal biogenesis factor 5-like a |
| chr4_-_39448829 | 1.33 |
ENSDART00000168526
|
si:dkey-261o4.9
|
si:dkey-261o4.9 |
| chr1_+_54124209 | 1.30 |
ENSDART00000187730
|
LO017722.1
|
|
| chr7_+_20393386 | 1.29 |
ENSDART00000173471
|
si:dkey-33c9.6
|
si:dkey-33c9.6 |
| chr22_+_2510828 | 1.28 |
ENSDART00000115348
ENSDART00000145611 |
zgc:173726
|
zgc:173726 |
| chr1_-_5745420 | 1.25 |
ENSDART00000166779
|
nrp2a
|
neuropilin 2a |
| chr4_+_54645654 | 1.20 |
ENSDART00000192864
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr13_+_25199849 | 1.17 |
ENSDART00000139209
ENSDART00000130876 |
ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr22_-_9973918 | 1.16 |
ENSDART00000146598
ENSDART00000106026 |
nlrc7
|
NLR family CARD domain containing 7 |
| chr3_+_15394428 | 1.16 |
ENSDART00000133168
|
atxn2l
|
ataxin 2-like |
| chr13_-_33153889 | 1.16 |
ENSDART00000141451
|
TC2N
|
tandem C2 domains, nuclear |
| chr24_-_37225877 | 1.16 |
ENSDART00000087438
|
cog7
|
component of oligomeric golgi complex 7 |
| chr22_-_10354381 | 1.15 |
ENSDART00000092050
|
stab1
|
stabilin 1 |
| chr2_-_9748039 | 1.11 |
ENSDART00000134870
|
si:ch1073-170o4.1
|
si:ch1073-170o4.1 |
| chr16_-_42175617 | 1.10 |
ENSDART00000084715
|
alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr12_+_22628704 | 1.09 |
ENSDART00000168935
|
mfsd8
|
major facilitator superfamily domain containing 8 |
| chr12_+_46608361 | 1.09 |
ENSDART00000189164
|
FADS6
|
fatty acid desaturase 6 |
| chr9_-_8670158 | 1.09 |
ENSDART00000077296
|
col4a1
|
collagen, type IV, alpha 1 |
| chr13_-_35459928 | 1.08 |
ENSDART00000144109
|
slx4ip
|
SLX4 interacting protein |
| chr21_-_2621127 | 1.06 |
ENSDART00000172363
|
col4a3bpb
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein b |
| chr2_+_38370535 | 1.06 |
ENSDART00000179614
|
psmb11a
|
proteasome subunit beta 11a |
| chr20_-_23946296 | 1.03 |
ENSDART00000143005
|
mdn1
|
midasin AAA ATPase 1 |
| chr24_-_30843250 | 0.99 |
ENSDART00000162920
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr19_+_170705 | 0.97 |
ENSDART00000053622
|
rnf139
|
ring finger protein 139 |
| chr2_-_24554416 | 0.96 |
ENSDART00000052061
|
cnn2
|
calponin 2 |
| chr4_-_57340828 | 0.95 |
ENSDART00000193689
|
zgc:194906
|
zgc:194906 |
| chr6_-_33925381 | 0.93 |
ENSDART00000137268
ENSDART00000145019 ENSDART00000141822 |
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr10_-_300000 | 0.93 |
ENSDART00000183273
|
emsy
|
EMSY BRCA2-interacting transcriptional repressor |
| chr2_+_9755422 | 0.91 |
ENSDART00000018524
|
pcyt1aa
|
phosphate cytidylyltransferase 1, choline, alpha a |
| chr24_+_1023839 | 0.88 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr21_+_38745094 | 0.84 |
ENSDART00000113316
|
heatr6
|
HEAT repeat containing 6 |
| chr3_+_634682 | 0.84 |
ENSDART00000164305
|
dicp1.1
|
diverse immunoglobulin domain-containing protein 1.1 |
| chr6_-_48087152 | 0.84 |
ENSDART00000180614
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of mafa+mafbb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 21.3 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 4.9 | 19.4 | GO:0019626 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 3.5 | 21.2 | GO:0050764 | regulation of phagocytosis(GO:0050764) |
| 3.4 | 10.2 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 2.5 | 14.8 | GO:0044209 | AMP salvage(GO:0044209) |
| 2.2 | 6.6 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 2.1 | 8.3 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 1.5 | 30.3 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 1.5 | 4.5 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 1.4 | 7.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 1.4 | 9.7 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 1.2 | 4.8 | GO:0031584 | activation of phospholipase D activity(GO:0031584) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 1.1 | 41.4 | GO:0032355 | response to estradiol(GO:0032355) |
| 1.1 | 4.3 | GO:0015840 | urea transport(GO:0015840) |
| 1.0 | 2.9 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 1.0 | 3.9 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.9 | 3.7 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.8 | 39.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.7 | 2.9 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.7 | 2.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.7 | 6.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.6 | 3.8 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.5 | 4.4 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.5 | 4.1 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.5 | 2.0 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.5 | 1.4 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.4 | 11.8 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.4 | 1.5 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.3 | 0.9 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.3 | 1.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.3 | 1.7 | GO:0008206 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.3 | 8.4 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.3 | 2.6 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.3 | 2.8 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.3 | 10.9 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.2 | 1.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.2 | 3.6 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 5.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.2 | 0.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 2.2 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.1 | 7.3 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 3.6 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.1 | 1.3 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 10.0 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 6.0 | GO:0007259 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.1 | 3.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 13.9 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 1.5 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.1 | 1.7 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.1 | 1.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 2.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 5.3 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 4.0 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.1 | 0.5 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.1 | 0.5 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.1 | 1.8 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 1.0 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.1 | 3.3 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 1.9 | GO:0032496 | response to lipopolysaccharide(GO:0032496) |
| 0.1 | 7.5 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.1 | 4.2 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.1 | 1.5 | GO:0042098 | T cell proliferation(GO:0042098) regulation of T cell proliferation(GO:0042129) |
| 0.1 | 0.2 | GO:0097240 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 2.0 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 1.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 7.7 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 1.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 3.4 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 5.1 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.4 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.0 | 0.5 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 2.4 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.1 | GO:1904182 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.0 | 2.6 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 8.2 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 2.9 | GO:0009063 | cellular amino acid catabolic process(GO:0009063) |
| 0.0 | 3.6 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 0.7 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.8 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 2.8 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 1.4 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 1.0 | GO:0036503 | ERAD pathway(GO:0036503) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.8 | 8.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.7 | 5.7 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.7 | 2.0 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.5 | 4.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 7.7 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 9.7 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.2 | 3.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 2.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 4.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 66.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 37.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 6.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 10.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 1.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 9.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 1.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 3.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 3.7 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 6.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 2.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 5.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 2.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 7.5 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.8 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 5.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 3.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 3.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 13.4 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.8 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 3.5 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 19.6 | GO:0005576 | extracellular region(GO:0005576) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.5 | 59.6 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 3.8 | 33.9 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 3.2 | 22.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 3.0 | 14.8 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 1.8 | 7.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 1.7 | 6.6 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 1.6 | 4.8 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 1.5 | 4.5 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 1.4 | 4.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 1.2 | 3.6 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.8 | 9.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.8 | 3.9 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.8 | 3.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.7 | 9.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.5 | 19.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.4 | 12.8 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.4 | 8.9 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.4 | 3.3 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.4 | 1.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 3.2 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.3 | 36.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 5.1 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.3 | 1.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.3 | 7.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 10.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.2 | 1.8 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.2 | 1.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.2 | 4.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.2 | 5.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 6.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 2.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 2.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 3.6 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 1.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 2.0 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 8.4 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 1.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 24.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 5.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 9.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 10.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 3.7 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 1.8 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 2.0 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.5 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 5.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 8.4 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.1 | 5.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 13.4 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 3.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 3.0 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 1.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 2.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 4.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 3.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 4.4 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
| 0.0 | 1.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 3.0 | GO:0016798 | hydrolase activity, acting on glycosyl bonds(GO:0016798) |
| 0.0 | 3.6 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.3 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 3.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 15.5 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 1.4 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.0 | 3.6 | GO:0008289 | lipid binding(GO:0008289) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 10.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.5 | 21.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.4 | 11.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.3 | 9.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.3 | 5.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.3 | 8.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.3 | 11.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.3 | 1.4 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.2 | 8.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.2 | 15.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 1.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 22.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 4.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 2.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 7.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 21.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 1.0 | 5.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.8 | 19.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.8 | 8.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.7 | 4.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.5 | 3.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.4 | 1.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.4 | 8.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.4 | 4.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.4 | 5.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 5.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.2 | 8.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 1.5 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 4.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.2 | 3.0 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 6.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 9.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 2.1 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |