Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
Results for klf12b
Z-value: 2.10
Transcription factors associated with klf12b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf12b
|
ENSDARG00000032197 | Kruppel-like factor 12b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf12b | dr11_v1_chr9_+_30720048_30720048 | 0.62 | 3.5e-11 | Click! |
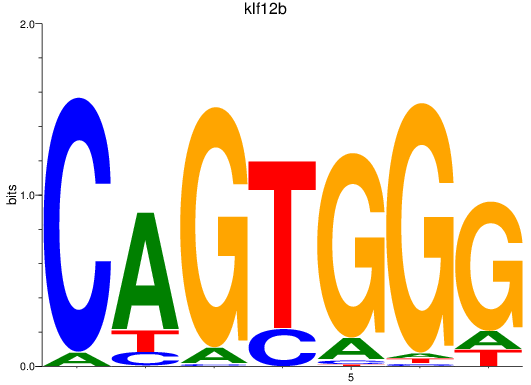
Activity profile of klf12b motif
Sorted Z-values of klf12b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_13742899 | 57.67 |
ENSDART00000104722
|
cdk5r2a
|
cyclin-dependent kinase 5, regulatory subunit 2a (p39) |
| chr25_-_4146947 | 30.58 |
ENSDART00000129268
|
fads2
|
fatty acid desaturase 2 |
| chr9_-_31278048 | 27.34 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr2_-_21082695 | 26.47 |
ENSDART00000032502
|
nebl
|
nebulette |
| chr11_-_37509001 | 24.17 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr2_+_47582681 | 22.57 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr8_+_31248917 | 20.76 |
ENSDART00000112170
|
unm_hu7912
|
un-named hu7912 |
| chr15_+_37197494 | 19.87 |
ENSDART00000166203
|
aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr21_-_43949208 | 19.72 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr18_+_18104235 | 19.66 |
ENSDART00000145342
|
cbln1
|
cerebellin 1 precursor |
| chr12_+_24344963 | 19.38 |
ENSDART00000191648
ENSDART00000183180 ENSDART00000088178 ENSDART00000189696 |
nrxn1a
|
neurexin 1a |
| chr7_-_18601206 | 18.82 |
ENSDART00000111636
|
DTX4
|
si:ch211-119e14.2 |
| chr20_-_39219537 | 18.42 |
ENSDART00000005764
|
cyp39a1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr5_-_55395964 | 18.18 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr8_+_31119548 | 18.17 |
ENSDART00000136578
|
syn1
|
synapsin I |
| chr5_-_22501663 | 18.11 |
ENSDART00000133174
|
si:dkey-27p18.5
|
si:dkey-27p18.5 |
| chr7_-_30082931 | 18.06 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr2_+_24203229 | 17.56 |
ENSDART00000138088
|
map4l
|
microtubule associated protein 4 like |
| chr17_+_15433518 | 17.53 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr1_-_21483832 | 17.33 |
ENSDART00000102790
|
glrba
|
glycine receptor, beta a |
| chr25_-_13842618 | 17.26 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr8_-_14050758 | 17.24 |
ENSDART00000133922
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr20_+_18580176 | 17.10 |
ENSDART00000185310
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr6_+_27667359 | 16.79 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr12_+_16233077 | 16.60 |
ENSDART00000152409
|
mpp3b
|
membrane protein, palmitoylated 3b (MAGUK p55 subfamily member 3) |
| chr17_-_52643970 | 16.53 |
ENSDART00000190594
|
spred1
|
sprouty-related, EVH1 domain containing 1 |
| chr12_+_9817440 | 16.23 |
ENSDART00000137081
ENSDART00000123712 |
rundc3ab
|
RUN domain containing 3Ab |
| chr16_-_12173554 | 16.04 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr2_-_14390627 | 15.90 |
ENSDART00000172367
|
sgip1b
|
SH3-domain GRB2-like (endophilin) interacting protein 1b |
| chr5_-_23317477 | 15.90 |
ENSDART00000090171
|
nlgn3b
|
neuroligin 3b |
| chr1_+_14283692 | 15.71 |
ENSDART00000017679
|
ppp2r2ca
|
protein phosphatase 2, regulatory subunit B, gamma a |
| chr6_+_24817852 | 15.41 |
ENSDART00000165609
|
barhl2
|
BarH-like homeobox 2 |
| chr8_-_7232413 | 15.21 |
ENSDART00000092426
|
grip2a
|
glutamate receptor interacting protein 2a |
| chr9_-_32753535 | 15.04 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr5_-_25174420 | 14.76 |
ENSDART00000141554
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr10_-_20637098 | 14.47 |
ENSDART00000080391
|
sprn2
|
shadow of prion protein 2 |
| chr19_-_13733870 | 14.36 |
ENSDART00000177773
|
epb41a
|
erythrocyte membrane protein band 4.1a |
| chr25_+_12640211 | 14.07 |
ENSDART00000165108
|
jph3
|
junctophilin 3 |
| chr6_-_30210378 | 14.00 |
ENSDART00000157359
ENSDART00000113924 |
lrrc7
|
leucine rich repeat containing 7 |
| chr11_+_36989696 | 13.98 |
ENSDART00000045888
|
tkta
|
transketolase a |
| chr1_+_38776294 | 13.89 |
ENSDART00000170546
|
wdr17
|
WD repeat domain 17 |
| chr2_+_22694382 | 13.87 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr3_-_18710009 | 13.86 |
ENSDART00000142478
|
grid2ipa
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, a |
| chr23_+_16633951 | 13.70 |
ENSDART00000109537
ENSDART00000193323 |
snphb
|
syntaphilin b |
| chr15_-_12319065 | 13.60 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr3_-_32320537 | 13.45 |
ENSDART00000113550
ENSDART00000168483 |
si:dkey-16p21.7
|
si:dkey-16p21.7 |
| chr4_+_6643421 | 13.28 |
ENSDART00000099462
|
gpr85
|
G protein-coupled receptor 85 |
| chr7_+_26224211 | 13.19 |
ENSDART00000173999
|
vgf
|
VGF nerve growth factor inducible |
| chr8_+_28900689 | 13.12 |
ENSDART00000141634
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr23_-_12345764 | 13.05 |
ENSDART00000133956
|
phactr3a
|
phosphatase and actin regulator 3a |
| chr16_+_20161805 | 12.80 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr25_-_19090479 | 12.56 |
ENSDART00000027465
ENSDART00000177670 |
cacna2d4b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4b |
| chr11_+_23957440 | 12.39 |
ENSDART00000190721
|
cntn2
|
contactin 2 |
| chr23_-_8373676 | 12.32 |
ENSDART00000105135
ENSDART00000158531 |
oprl1
|
opiate receptor-like 1 |
| chr3_-_19091024 | 12.18 |
ENSDART00000188485
ENSDART00000110554 |
grin2ca
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca |
| chr16_-_29458806 | 12.11 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr17_-_36896560 | 11.87 |
ENSDART00000045287
|
mapre3a
|
microtubule-associated protein, RP/EB family, member 3a |
| chr7_-_19940473 | 11.84 |
ENSDART00000127669
|
prox1b
|
prospero homeobox 1b |
| chr20_+_5564042 | 11.82 |
ENSDART00000090934
ENSDART00000127050 |
nrxn3b
|
neurexin 3b |
| chr25_+_19955598 | 11.71 |
ENSDART00000091547
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr14_+_32022272 | 11.66 |
ENSDART00000105760
|
zic6
|
zic family member 6 |
| chr7_-_43840680 | 11.59 |
ENSDART00000002279
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr14_-_27121854 | 11.57 |
ENSDART00000173119
|
pcdh11
|
protocadherin 11 |
| chr22_+_5103349 | 11.48 |
ENSDART00000083474
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr2_+_27010439 | 11.45 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr17_+_18117029 | 11.43 |
ENSDART00000154646
ENSDART00000179739 |
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr15_+_32821392 | 11.34 |
ENSDART00000158272
|
dclk1b
|
doublecortin-like kinase 1b |
| chr8_+_36803415 | 11.29 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr1_+_20084389 | 11.13 |
ENSDART00000140263
|
prss12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr3_-_28750495 | 11.12 |
ENSDART00000054408
|
gsg1l
|
gsg1-like |
| chr10_+_5159475 | 11.12 |
ENSDART00000142507
|
cdc42se2
|
CDC42 small effector 2 |
| chr20_-_19422496 | 11.09 |
ENSDART00000143658
|
si:ch211-278j3.3
|
si:ch211-278j3.3 |
| chr9_-_35155089 | 11.06 |
ENSDART00000077901
|
appb
|
amyloid beta (A4) precursor protein b |
| chr10_+_29698467 | 11.06 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr4_+_19535946 | 10.93 |
ENSDART00000192342
ENSDART00000183740 ENSDART00000180812 ENSDART00000180017 |
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr17_+_18117358 | 10.87 |
ENSDART00000144894
|
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr9_-_27442339 | 10.79 |
ENSDART00000138602
|
stxbp5l
|
syntaxin binding protein 5-like |
| chr10_+_21776911 | 10.69 |
ENSDART00000163077
ENSDART00000186093 |
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr20_-_47704973 | 10.68 |
ENSDART00000174808
|
tfap2b
|
transcription factor AP-2 beta |
| chr4_-_1324141 | 10.65 |
ENSDART00000180720
|
ptn
|
pleiotrophin |
| chr19_-_26863626 | 10.43 |
ENSDART00000145568
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr3_-_38692920 | 10.36 |
ENSDART00000155042
|
mpp3a
|
membrane protein, palmitoylated 3a (MAGUK p55 subfamily member 3) |
| chr10_+_22381802 | 10.17 |
ENSDART00000112484
|
nlgn2b
|
neuroligin 2b |
| chr22_-_26323893 | 10.16 |
ENSDART00000105099
|
capn1b
|
calpain 1, (mu/I) large subunit b |
| chr24_-_21989406 | 10.15 |
ENSDART00000032963
|
apoob
|
apolipoprotein O, b |
| chr25_+_37126921 | 10.11 |
ENSDART00000124331
|
si:ch1073-174d20.1
|
si:ch1073-174d20.1 |
| chr11_+_23933016 | 10.04 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr24_+_32472155 | 10.02 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr20_+_17739923 | 9.89 |
ENSDART00000024627
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr7_-_57933736 | 9.89 |
ENSDART00000142580
|
ank2b
|
ankyrin 2b, neuronal |
| chr23_+_35714574 | 9.79 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr5_+_20148671 | 9.63 |
ENSDART00000143205
|
svopa
|
SV2 related protein a |
| chr4_+_5741733 | 9.61 |
ENSDART00000110243
|
pou3f2a
|
POU class 3 homeobox 2a |
| chr7_+_33314925 | 9.61 |
ENSDART00000148590
|
coro2ba
|
coronin, actin binding protein, 2Ba |
| chr4_+_8168514 | 9.52 |
ENSDART00000150830
|
ninj2
|
ninjurin 2 |
| chr18_-_16179129 | 9.44 |
ENSDART00000125353
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr3_-_37476475 | 9.43 |
ENSDART00000148107
|
si:ch211-278a6.1
|
si:ch211-278a6.1 |
| chr19_-_43552252 | 9.42 |
ENSDART00000138308
|
gpr186
|
G protein-coupled receptor 186 |
| chr8_-_12468744 | 9.40 |
ENSDART00000135019
|
FIBCD1 (1 of many)
|
si:dkeyp-51b7.3 |
| chr17_-_3303805 | 9.33 |
ENSDART00000169136
|
CABZ01007222.1
|
|
| chr24_-_10828560 | 9.31 |
ENSDART00000132282
|
fam49bb
|
family with sequence similarity 49, member Bb |
| chr15_-_11341635 | 9.28 |
ENSDART00000055220
|
rab30
|
RAB30, member RAS oncogene family |
| chr15_-_8517555 | 9.19 |
ENSDART00000140213
|
npas1
|
neuronal PAS domain protein 1 |
| chr7_-_34413392 | 9.14 |
ENSDART00000173673
ENSDART00000047208 ENSDART00000174546 |
madd
|
MAP-kinase activating death domain |
| chr23_-_24450686 | 9.09 |
ENSDART00000189161
|
spen
|
spen family transcriptional repressor |
| chr4_-_28158335 | 9.00 |
ENSDART00000134605
|
gramd4a
|
GRAM domain containing 4a |
| chr12_+_2381213 | 8.98 |
ENSDART00000188007
|
LO018238.1
|
|
| chr3_-_19133003 | 8.93 |
ENSDART00000145215
|
grin2ca
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca |
| chr21_-_24632778 | 8.92 |
ENSDART00000132533
ENSDART00000058370 |
arhgap32b
|
Rho GTPase activating protein 32b |
| chr14_-_2209742 | 8.88 |
ENSDART00000054889
|
pcdh2ab5
|
protocadherin 2 alpha b 5 |
| chr24_-_11325849 | 8.86 |
ENSDART00000182485
|
myrip
|
myosin VIIA and Rab interacting protein |
| chr2_+_33368414 | 8.81 |
ENSDART00000077462
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr1_-_57172294 | 8.71 |
ENSDART00000063774
|
rac1l
|
Rac family small GTPase 1, like |
| chr8_+_26868105 | 8.70 |
ENSDART00000005337
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr20_+_38201644 | 8.70 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr12_+_21299338 | 8.69 |
ENSDART00000074540
ENSDART00000133188 |
ca10a
|
carbonic anhydrase Xa |
| chr4_-_23643272 | 8.53 |
ENSDART00000112301
ENSDART00000133184 |
trhde.2
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 2 |
| chr19_-_44089509 | 8.52 |
ENSDART00000189136
|
rad21b
|
RAD21 cohesin complex component b |
| chr5_-_51903243 | 8.52 |
ENSDART00000125535
|
mtx3
|
metaxin 3 |
| chr13_-_31470439 | 8.49 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr15_+_29472065 | 8.47 |
ENSDART00000154343
|
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr5_-_31897139 | 8.47 |
ENSDART00000191957
|
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr3_+_32391540 | 8.38 |
ENSDART00000156608
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr4_-_18201622 | 8.23 |
ENSDART00000133509
|
anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr21_-_12272543 | 8.22 |
ENSDART00000081510
ENSDART00000151297 |
celf4
|
CUGBP, Elav-like family member 4 |
| chr22_+_12431608 | 8.11 |
ENSDART00000108609
|
rnd3a
|
Rho family GTPase 3a |
| chr22_-_600016 | 8.08 |
ENSDART00000086434
|
tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr5_-_21970881 | 8.07 |
ENSDART00000182907
|
arhgef9a
|
Cdc42 guanine nucleotide exchange factor (GEF) 9a |
| chr13_+_4505232 | 8.07 |
ENSDART00000007500
ENSDART00000161684 |
pde10a
|
phosphodiesterase 10A |
| chr5_-_38506981 | 8.07 |
ENSDART00000097822
|
atp1b2b
|
ATPase Na+/K+ transporting subunit beta 2b |
| chr15_+_40188076 | 8.06 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr18_+_18612388 | 8.02 |
ENSDART00000186455
|
st3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr23_-_24483311 | 7.99 |
ENSDART00000185793
ENSDART00000109248 |
spen
|
spen family transcriptional repressor |
| chr16_+_37470717 | 7.93 |
ENSDART00000112003
ENSDART00000188431 ENSDART00000192837 |
adgrb1a
|
adhesion G protein-coupled receptor B1a |
| chr15_+_47418565 | 7.87 |
ENSDART00000155709
|
clpb
|
ClpB homolog, mitochondrial AAA ATPase chaperonin |
| chr2_+_31833997 | 7.86 |
ENSDART00000066788
|
epdr1
|
ependymin related 1 |
| chr14_-_44841503 | 7.85 |
ENSDART00000179114
|
GRXCR1
|
si:dkey-109l4.6 |
| chr18_-_14937211 | 7.78 |
ENSDART00000141893
|
mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr21_+_26748141 | 7.75 |
ENSDART00000169025
|
pcxa
|
pyruvate carboxylase a |
| chr13_+_19322686 | 7.75 |
ENSDART00000058036
|
emx2
|
empty spiracles homeobox 2 |
| chr10_+_25222367 | 7.62 |
ENSDART00000042767
|
grm5a
|
glutamate receptor, metabotropic 5a |
| chr3_+_54047342 | 7.62 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr2_+_50094873 | 7.57 |
ENSDART00000132307
|
zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr22_-_22130623 | 7.49 |
ENSDART00000113168
|
CU855878.1
|
|
| chr14_-_2213660 | 7.48 |
ENSDART00000162537
|
pcdh2ab3
|
protocadherin 2 alpha b 3 |
| chr12_+_42574148 | 7.40 |
ENSDART00000157855
|
ebf3a
|
early B cell factor 3a |
| chr3_-_22829710 | 7.36 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr20_+_26349002 | 7.34 |
ENSDART00000152842
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr6_-_20875111 | 7.33 |
ENSDART00000115118
ENSDART00000159916 |
tns1a
|
tensin 1a |
| chr9_+_40939336 | 7.31 |
ENSDART00000100386
|
mstnb
|
myostatin b |
| chr2_-_36818132 | 7.27 |
ENSDART00000110447
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr11_+_14004236 | 7.26 |
ENSDART00000162478
ENSDART00000171076 |
grin3bb
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3Bb |
| chr15_-_20024205 | 7.24 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr23_+_16638639 | 7.13 |
ENSDART00000143545
|
snphb
|
syntaphilin b |
| chr16_-_44512882 | 7.07 |
ENSDART00000191241
|
CR925804.2
|
|
| chr7_+_61906903 | 7.00 |
ENSDART00000108540
|
tdrd7b
|
tudor domain containing 7 b |
| chr24_+_24461558 | 6.99 |
ENSDART00000182424
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr1_+_29183962 | 6.93 |
ENSDART00000113735
|
cars2
|
cysteinyl-tRNA synthetase 2, mitochondrial |
| chr24_+_29382109 | 6.88 |
ENSDART00000184620
ENSDART00000188414 ENSDART00000186132 ENSDART00000191489 |
ntng1a
|
netrin g1a |
| chr19_+_27479838 | 6.86 |
ENSDART00000103922
|
atat1
|
alpha tubulin acetyltransferase 1 |
| chr4_-_76484737 | 6.84 |
ENSDART00000183816
|
ftr51
|
finTRIM family, member 51 |
| chr5_+_33301005 | 6.75 |
ENSDART00000006021
|
usp20
|
ubiquitin specific peptidase 20 |
| chr20_-_23226453 | 6.69 |
ENSDART00000142721
|
dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr20_+_14977260 | 6.68 |
ENSDART00000186424
|
vamp4
|
vesicle-associated membrane protein 4 |
| chr1_+_1599979 | 6.67 |
ENSDART00000097626
|
urp2
|
urotensin II-related peptide |
| chr17_-_12758171 | 6.66 |
ENSDART00000131564
|
brms1la
|
breast cancer metastasis-suppressor 1-like a |
| chr17_-_8692722 | 6.53 |
ENSDART00000148931
ENSDART00000192891 |
ctbp2a
|
C-terminal binding protein 2a |
| chr24_-_10919588 | 6.50 |
ENSDART00000131204
|
asap1b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1b |
| chr14_-_39074539 | 6.50 |
ENSDART00000030509
|
glra4a
|
glycine receptor, alpha 4a |
| chr8_+_42998944 | 6.45 |
ENSDART00000048819
|
rassf2a
|
Ras association (RalGDS/AF-6) domain family member 2a |
| chr6_-_7842078 | 6.45 |
ENSDART00000065507
|
plppr2b
|
phospholipid phosphatase related 2b |
| chr4_+_23223881 | 6.44 |
ENSDART00000133056
ENSDART00000089126 |
trhde.1
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 1 |
| chr16_-_44649053 | 6.41 |
ENSDART00000184807
|
CR925804.2
|
|
| chr19_-_3488860 | 6.39 |
ENSDART00000172520
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr5_+_52039067 | 6.33 |
ENSDART00000143276
|
setbp1
|
SET binding protein 1 |
| chr8_-_53108207 | 6.29 |
ENSDART00000111023
|
b3galt4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 |
| chr11_-_42554290 | 6.24 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr14_+_28281744 | 6.21 |
ENSDART00000173292
|
mid2
|
midline 2 |
| chr16_+_14812585 | 6.20 |
ENSDART00000134087
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr18_-_26675699 | 6.10 |
ENSDART00000113280
|
FRMD5
|
si:ch211-69m14.1 |
| chr5_+_22098591 | 6.00 |
ENSDART00000143676
|
zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr8_+_25247245 | 5.98 |
ENSDART00000045798
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr15_+_1397811 | 5.96 |
ENSDART00000102125
|
schip1
|
schwannomin interacting protein 1 |
| chr24_+_26134029 | 5.96 |
ENSDART00000185134
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr18_-_41211980 | 5.93 |
ENSDART00000171431
|
zbtb38
|
zinc finger and BTB domain containing 38 |
| chr14_-_2322484 | 5.89 |
ENSDART00000167806
|
si:ch73-379j16.2
|
si:ch73-379j16.2 |
| chr23_+_19590598 | 5.87 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr23_+_11669337 | 5.84 |
ENSDART00000131355
|
cntn3a.1
|
contactin 3a, tandem duplicate 1 |
| chr7_+_44713135 | 5.84 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr23_+_11669109 | 5.84 |
ENSDART00000091416
|
cntn3a.1
|
contactin 3a, tandem duplicate 1 |
| chr17_+_16564921 | 5.82 |
ENSDART00000151904
|
foxn3
|
forkhead box N3 |
| chr15_+_34062460 | 5.82 |
ENSDART00000164654
|
si:dkey-30e9.6
|
si:dkey-30e9.6 |
| chr20_-_31781941 | 5.77 |
ENSDART00000139417
|
stxbp5a
|
syntaxin binding protein 5a (tomosyn) |
| chr18_-_15373620 | 5.76 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr8_+_47342586 | 5.73 |
ENSDART00000007624
|
plch2a
|
phospholipase C, eta 2a |
| chr5_+_11812089 | 5.71 |
ENSDART00000111359
|
fbxo21
|
F-box protein 21 |
| chr25_+_8662469 | 5.68 |
ENSDART00000154680
ENSDART00000188568 |
man2a2
|
mannosidase, alpha, class 2A, member 2 |
| chr3_-_17871846 | 5.67 |
ENSDART00000074478
ENSDART00000187941 |
nkiras2
|
NFKB inhibitor interacting Ras-like 2 |
| chr13_+_4505079 | 5.63 |
ENSDART00000144312
|
pde10a
|
phosphodiesterase 10A |
| chr19_+_19028633 | 5.61 |
ENSDART00000158618
|
cpne4b
|
copine IVb |
Network of associatons between targets according to the STRING database.
First level regulatory network of klf12b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 22.4 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 4.2 | 20.8 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 3.9 | 27.3 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 3.8 | 15.0 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 3.4 | 20.4 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 3.3 | 9.9 | GO:0021577 | hindbrain structural organization(GO:0021577) dorsal fin morphogenesis(GO:0035142) cell motility involved in somitogenic axis elongation(GO:0090247) cell migration involved in somitogenic axis elongation(GO:0090248) |
| 3.0 | 33.3 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 2.8 | 14.0 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 2.7 | 13.7 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 2.5 | 7.6 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 2.4 | 24.2 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 2.4 | 26.1 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 2.4 | 9.4 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 2.3 | 20.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 2.3 | 9.0 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 2.2 | 6.7 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 2.2 | 8.8 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 2.2 | 23.8 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 2.2 | 10.8 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 2.0 | 9.9 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 1.8 | 7.3 | GO:0045843 | negative regulation of striated muscle tissue development(GO:0045843) negative regulation of muscle tissue development(GO:1901862) |
| 1.7 | 6.9 | GO:0021828 | pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 1.7 | 6.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 1.6 | 15.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 1.4 | 4.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 1.3 | 11.9 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 1.3 | 49.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 1.3 | 3.8 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 1.2 | 23.6 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 1.1 | 4.6 | GO:0045730 | respiratory burst(GO:0045730) |
| 1.1 | 3.4 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 1.1 | 20.4 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 1.1 | 9.5 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 1.0 | 14.0 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 1.0 | 6.0 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 1.0 | 7.8 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 1.0 | 7.7 | GO:0060118 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.9 | 12.3 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.9 | 5.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.9 | 4.6 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.9 | 3.7 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.9 | 8.5 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.8 | 12.6 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.8 | 29.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.8 | 5.0 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.8 | 4.1 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.8 | 4.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.8 | 15.2 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.8 | 9.9 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.7 | 16.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.7 | 7.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.7 | 7.8 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.7 | 10.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.7 | 21.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.7 | 8.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.6 | 3.8 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.6 | 17.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.6 | 3.8 | GO:0006699 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.6 | 8.1 | GO:0001964 | startle response(GO:0001964) |
| 0.6 | 2.3 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.6 | 22.6 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.5 | 26.9 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.5 | 17.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.5 | 3.2 | GO:0043091 | amino acid import(GO:0043090) L-arginine import(GO:0043091) L-amino acid import(GO:0043092) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.5 | 16.8 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.5 | 8.1 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.5 | 29.2 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.5 | 1.4 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.5 | 8.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.4 | 2.6 | GO:0008584 | male gonad development(GO:0008584) |
| 0.4 | 6.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.4 | 16.6 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.4 | 11.7 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.4 | 9.3 | GO:0050870 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.4 | 21.4 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.4 | 3.8 | GO:2000144 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.4 | 9.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.3 | 2.4 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.3 | 2.0 | GO:0042670 | retinal cone cell differentiation(GO:0042670) |
| 0.3 | 2.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.3 | 5.4 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.3 | 1.6 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.3 | 3.2 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.3 | 14.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.3 | 2.2 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.3 | 4.9 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.3 | 2.7 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.3 | 5.0 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.3 | 1.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 | 12.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 5.8 | GO:0007418 | ventral midline development(GO:0007418) |
| 0.3 | 2.3 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.3 | 8.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.3 | 28.0 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.3 | 2.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.3 | 6.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.3 | 3.1 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 1.1 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.3 | 3.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.3 | 1.9 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.3 | 5.6 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.3 | 6.0 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.2 | 48.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 0.7 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.2 | 2.1 | GO:0045738 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.2 | 12.3 | GO:0003146 | heart jogging(GO:0003146) |
| 0.2 | 2.0 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 1.1 | GO:0072574 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.2 | 10.0 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.2 | 13.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.2 | 4.3 | GO:0007020 | microtubule nucleation(GO:0007020) microtubule anchoring(GO:0034453) |
| 0.2 | 11.1 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.2 | 1.4 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.2 | 8.4 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 1.2 | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902229) |
| 0.2 | 1.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 1.9 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.2 | 2.5 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 0.6 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.2 | 1.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 | 0.9 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.2 | 5.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.2 | 3.9 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.2 | 9.6 | GO:0003401 | axis elongation(GO:0003401) |
| 0.2 | 3.3 | GO:0043489 | RNA stabilization(GO:0043489) |
| 0.2 | 2.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 2.9 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.2 | 21.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.2 | 1.0 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.2 | 10.4 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.2 | 2.7 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.2 | 1.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.2 | 3.1 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.1 | 1.6 | GO:0021537 | telencephalon development(GO:0021537) |
| 0.1 | 17.9 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.1 | 1.4 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 3.6 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 1.8 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 3.9 | GO:0006505 | GPI anchor metabolic process(GO:0006505) |
| 0.1 | 7.8 | GO:0008593 | regulation of Notch signaling pathway(GO:0008593) |
| 0.1 | 2.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.0 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 5.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 15.3 | GO:0045216 | cell-cell junction organization(GO:0045216) |
| 0.1 | 6.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 1.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 2.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.2 | GO:0046379 | hyaluronan biosynthetic process(GO:0030213) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 10.9 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.1 | 0.3 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.1 | 7.4 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.1 | 2.5 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 0.8 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 1.6 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 3.6 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 1.1 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.1 | 0.2 | GO:2000105 | positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.1 | 2.2 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 1.9 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 3.4 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 5.7 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.1 | 0.6 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.1 | 1.5 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 6.2 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 0.9 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 4.5 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.1 | 0.8 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.1 | 3.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.6 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 2.1 | GO:0045727 | positive regulation of cellular amide metabolic process(GO:0034250) positive regulation of translation(GO:0045727) |
| 0.1 | 19.8 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.1 | 4.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 2.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 6.9 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.1 | 13.7 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 0.5 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 1.7 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 0.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 1.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 8.2 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.3 | GO:0034340 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.3 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 1.7 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.9 | GO:0042593 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.0 | 0.5 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 1.7 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.6 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 5.8 | GO:0007005 | mitochondrion organization(GO:0007005) |
| 0.0 | 0.7 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 1.4 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 3.3 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 1.3 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 2.0 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 0.2 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 1.2 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.5 | 57.7 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 3.4 | 10.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 2.4 | 24.2 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 1.9 | 11.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.7 | 20.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 1.7 | 24.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.4 | 13.9 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 1.3 | 8.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 1.2 | 8.5 | GO:0030892 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 1.1 | 8.5 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 1.0 | 6.2 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.9 | 10.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.9 | 11.7 | GO:0033270 | paranode region of axon(GO:0033270) juxtaparanode region of axon(GO:0044224) |
| 0.8 | 8.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.8 | 5.5 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.8 | 6.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.8 | 9.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.7 | 3.4 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.7 | 3.4 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.6 | 9.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.6 | 9.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.6 | 4.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.5 | 11.6 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.5 | 11.9 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.5 | 24.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.5 | 7.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.5 | 15.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.5 | 11.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.5 | 2.3 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.4 | 18.7 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.4 | 3.5 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.4 | 2.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.4 | 1.8 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.4 | 1.4 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.4 | 18.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.3 | 1.3 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.3 | 34.2 | GO:0099572 | postsynaptic specialization(GO:0099572) |
| 0.3 | 6.7 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.3 | 18.2 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 60.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.3 | 3.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 4.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 21.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.3 | 22.6 | GO:0030141 | secretory granule(GO:0030141) |
| 0.2 | 0.7 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.2 | 11.2 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.2 | 3.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 6.2 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 2.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 4.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 15.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 4.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 9.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 5.1 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.1 | 1.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 4.1 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 54.1 | GO:0043005 | neuron projection(GO:0043005) |
| 0.1 | 10.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 3.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 4.6 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 4.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 2.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 11.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 1.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 16.6 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 2.1 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 12.7 | GO:0035770 | ribonucleoprotein granule(GO:0035770) |
| 0.1 | 2.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 2.2 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 20.3 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 17.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 2.5 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 25.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 3.6 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 24.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 100.3 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 5.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.3 | GO:0030496 | midbody(GO:0030496) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 57.7 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 3.0 | 33.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 2.9 | 8.7 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 2.7 | 13.7 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 2.6 | 23.8 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 2.6 | 7.8 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 2.5 | 7.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 2.3 | 14.0 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 2.2 | 10.8 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 1.9 | 9.4 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 1.8 | 5.5 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 1.6 | 32.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 1.4 | 12.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 1.3 | 24.2 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 1.3 | 3.9 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 1.2 | 17.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 1.1 | 7.6 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 1.1 | 17.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 1.0 | 5.2 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 1.0 | 4.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 1.0 | 26.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 1.0 | 3.8 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.9 | 3.8 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.9 | 8.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.7 | 23.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.7 | 8.8 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.6 | 15.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.6 | 13.9 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.6 | 4.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.6 | 17.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.6 | 3.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.5 | 6.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.5 | 2.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.5 | 8.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.5 | 20.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.5 | 9.9 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.5 | 6.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.5 | 12.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.5 | 13.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.5 | 4.9 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.5 | 17.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.5 | 3.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.5 | 2.3 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.5 | 2.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.4 | 3.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.4 | 14.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 13.1 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.4 | 13.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.4 | 1.6 | GO:0051800 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.4 | 8.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.4 | 2.4 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.4 | 19.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.4 | 8.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.4 | 37.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.4 | 1.4 | GO:0045547 | dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.3 | 34.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.3 | 8.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 2.8 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.3 | 9.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.3 | 8.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.3 | 6.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.3 | 4.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 3.1 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 1.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.3 | 3.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 3.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 6.3 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.2 | 21.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.2 | 3.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 5.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 3.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 8.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 0.9 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.2 | 0.9 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.2 | 1.4 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 2.8 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 0.7 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.2 | 2.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 1.8 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.2 | 8.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 1.6 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 5.0 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.2 | 1.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 2.7 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 3.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 0.5 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.2 | 20.2 | GO:0042626 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.1 | 1.8 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 6.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 2.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 2.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 10.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 2.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 3.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.4 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 2.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 9.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 6.0 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.1 | 1.1 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.1 | 11.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 4.0 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.1 | 6.0 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 2.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 18.4 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.1 | 3.9 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.1 | 13.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 1.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 3.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 17.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 5.2 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.1 | 4.4 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.1 | 1.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 5.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 39.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.6 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 11.1 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 12.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.9 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 2.3 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 12.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.2 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 2.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.7 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.8 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 2.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 45.8 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 1.7 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
| 0.0 | 1.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 9.2 | GO:0016491 | oxidoreductase activity(GO:0016491) |
| 0.0 | 9.0 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.7 | 13.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.5 | 8.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.5 | 3.7 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.4 | 3.8 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.4 | 4.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.4 | 4.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.4 | 13.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.3 | 13.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.3 | 9.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.2 | 9.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 8.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 2.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 0.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 2.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 1.9 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 1.8 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 4.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.6 | 19.7 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 3.7 | 18.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 3.6 | 18.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 3.1 | 30.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 1.7 | 19.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 1.4 | 13.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 1.3 | 18.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.9 | 8.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.8 | 12.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.7 | 7.1 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.7 | 16.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.5 | 3.7 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.5 | 26.2 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.4 | 10.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.4 | 6.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.4 | 5.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.3 | 4.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 6.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.3 | 4.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.2 | 0.9 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 5.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 4.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 1.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 2.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 3.9 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 5.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.7 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 4.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 0.2 | REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | Genes involved in Downstream signaling of activated FGFR |
| 0.1 | 5.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 2.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 3.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.6 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |