Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
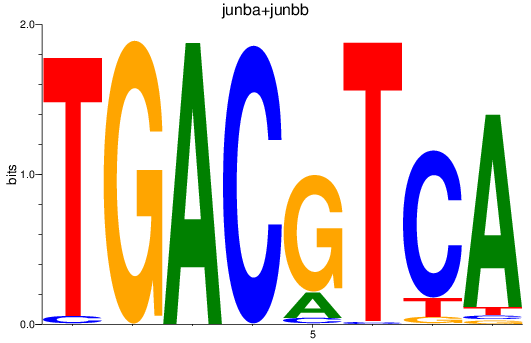
Results for junba+junbb
Z-value: 2.39
Transcription factors associated with junba+junbb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
junba
|
ENSDARG00000074378 | JunB proto-oncogene, AP-1 transcription factor subunit a |
|
junbb
|
ENSDARG00000104773 | JunB proto-oncogene, AP-1 transcription factor subunit b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| junba | dr11_v1_chr1_-_51734524_51734535 | -0.29 | 3.9e-03 | Click! |
| junbb | dr11_v1_chr3_-_7656059_7656059 | -0.05 | 6.2e-01 | Click! |
Activity profile of junba+junbb motif
Sorted Z-values of junba+junbb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_12385308 | 62.94 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr6_+_27667359 | 41.60 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr20_+_34913069 | 39.22 |
ENSDART00000007584
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr14_+_34486629 | 32.37 |
ENSDART00000131861
|
tmsb2
|
thymosin beta 2 |
| chr2_-_11512819 | 32.08 |
ENSDART00000142013
|
penka
|
proenkephalin a |
| chr16_-_17207754 | 29.24 |
ENSDART00000063804
|
wu:fj39g12
|
wu:fj39g12 |
| chr10_-_39011514 | 28.95 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr3_-_36115339 | 27.94 |
ENSDART00000187406
ENSDART00000123505 ENSDART00000151775 |
rab11fip4a
|
RAB11 family interacting protein 4 (class II) a |
| chr16_-_43026273 | 26.09 |
ENSDART00000156820
ENSDART00000189080 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr1_-_22861348 | 25.78 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr1_-_38756870 | 25.77 |
ENSDART00000130324
ENSDART00000148404 |
gpm6ab
|
glycoprotein M6Ab |
| chr19_+_24882845 | 25.03 |
ENSDART00000010580
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr8_+_39607466 | 24.91 |
ENSDART00000097427
|
msi1
|
musashi RNA-binding protein 1 |
| chr3_+_32403758 | 24.52 |
ENSDART00000156982
|
si:ch211-195b15.8
|
si:ch211-195b15.8 |
| chr2_-_42415902 | 24.36 |
ENSDART00000142489
|
slco5a1b
|
solute carrier organic anion transporter family member 5A1b |
| chr1_-_51606552 | 24.29 |
ENSDART00000130828
|
cnrip1a
|
cannabinoid receptor interacting protein 1a |
| chr3_-_49566364 | 23.82 |
ENSDART00000161507
|
zgc:153426
|
zgc:153426 |
| chr5_-_35301800 | 23.41 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr15_-_44512461 | 23.19 |
ENSDART00000155456
|
gria4a
|
glutamate receptor, ionotropic, AMPA 4a |
| chr18_-_38087875 | 22.72 |
ENSDART00000111301
|
luzp2
|
leucine zipper protein 2 |
| chr18_+_21408794 | 22.48 |
ENSDART00000140161
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr16_-_43025885 | 21.88 |
ENSDART00000193146
ENSDART00000157302 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr9_+_219124 | 21.87 |
ENSDART00000161484
|
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr23_+_19590006 | 21.73 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr19_+_10396042 | 21.26 |
ENSDART00000028048
ENSDART00000151735 |
necap1
|
NECAP endocytosis associated 1 |
| chr6_-_31348999 | 21.03 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr6_+_27146671 | 20.23 |
ENSDART00000156792
|
kif1aa
|
kinesin family member 1Aa |
| chr20_+_27020201 | 20.18 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr15_-_12319065 | 20.02 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr24_-_7632187 | 19.98 |
ENSDART00000041714
|
atp6v0a1b
|
ATPase H+ transporting V0 subunit a1b |
| chr3_-_13147310 | 19.84 |
ENSDART00000160840
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr11_+_25064519 | 19.80 |
ENSDART00000016181
|
ndrg3a
|
ndrg family member 3a |
| chr8_+_23165749 | 19.53 |
ENSDART00000063057
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr18_+_16330025 | 19.51 |
ENSDART00000142353
|
nts
|
neurotensin |
| chr2_+_22694382 | 19.37 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr20_-_34801181 | 19.36 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr24_-_17444067 | 19.27 |
ENSDART00000155843
|
cntnap2a
|
contactin associated protein like 2a |
| chr12_-_19103490 | 19.03 |
ENSDART00000060561
|
csdc2a
|
cold shock domain containing C2, RNA binding a |
| chr4_+_13733838 | 18.92 |
ENSDART00000067166
ENSDART00000133157 |
cntn1b
|
contactin 1b |
| chr12_+_45200744 | 18.82 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr8_+_8845932 | 18.75 |
ENSDART00000112028
|
si:ch211-180f4.1
|
si:ch211-180f4.1 |
| chr1_+_49266886 | 18.62 |
ENSDART00000137179
|
caly
|
calcyon neuron-specific vesicular protein |
| chr7_+_23907692 | 17.96 |
ENSDART00000045479
|
syt4
|
synaptotagmin IV |
| chr20_+_3108597 | 17.92 |
ENSDART00000133435
|
CEP170B (1 of many)
|
si:ch73-212j7.1 |
| chr11_-_7320211 | 17.44 |
ENSDART00000091664
|
apc2
|
adenomatosis polyposis coli 2 |
| chr5_+_3501859 | 16.61 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr19_+_16222618 | 16.49 |
ENSDART00000137189
ENSDART00000169246 ENSDART00000190583 ENSDART00000189521 |
ptprua
|
protein tyrosine phosphatase, receptor type, U, a |
| chr18_-_38088099 | 16.09 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr18_+_17663898 | 15.84 |
ENSDART00000021213
|
cpne2
|
copine II |
| chr21_+_22630297 | 15.78 |
ENSDART00000147175
|
si:dkeyp-69c1.7
|
si:dkeyp-69c1.7 |
| chr22_+_11535131 | 15.74 |
ENSDART00000113930
|
npb
|
neuropeptide B |
| chr21_+_22630627 | 15.74 |
ENSDART00000193092
|
si:dkeyp-69c1.7
|
si:dkeyp-69c1.7 |
| chr3_+_31933893 | 15.58 |
ENSDART00000146509
ENSDART00000139644 |
lin7b
|
lin-7 homolog B (C. elegans) |
| chr19_+_25649626 | 15.56 |
ENSDART00000146947
|
tac1
|
tachykinin 1 |
| chr25_-_7686201 | 15.54 |
ENSDART00000157267
ENSDART00000155094 |
si:ch211-286c4.6
|
si:ch211-286c4.6 |
| chr11_+_40649412 | 15.54 |
ENSDART00000043016
ENSDART00000134560 |
slc45a1
|
solute carrier family 45, member 1 |
| chr9_+_11532025 | 15.34 |
ENSDART00000109037
|
cdk5r2b
|
cyclin-dependent kinase 5, regulatory subunit 2b (p39) |
| chr17_+_24722646 | 15.33 |
ENSDART00000138356
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr24_-_28711176 | 15.21 |
ENSDART00000105753
|
olfm3a
|
olfactomedin 3a |
| chr10_+_15777258 | 15.07 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr7_-_72605673 | 15.03 |
ENSDART00000123887
|
MAPK8IP1 (1 of many)
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr24_+_34606966 | 14.77 |
ENSDART00000105477
|
lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr1_+_31864404 | 14.76 |
ENSDART00000075260
|
inab
|
internexin neuronal intermediate filament protein, alpha b |
| chr8_+_25959940 | 14.74 |
ENSDART00000143011
ENSDART00000140626 |
si:dkey-72l14.4
|
si:dkey-72l14.4 |
| chr15_-_28200049 | 14.50 |
ENSDART00000004200
|
sarm1
|
sterile alpha and TIR motif containing 1 |
| chr12_-_30443562 | 14.48 |
ENSDART00000020769
|
adrb1
|
adrenoceptor beta 1 |
| chr17_-_26721007 | 14.43 |
ENSDART00000034580
|
calm1a
|
calmodulin 1a |
| chr8_+_23174137 | 14.29 |
ENSDART00000189470
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr21_-_43606502 | 14.28 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr16_+_14029283 | 14.20 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr12_-_33972798 | 14.20 |
ENSDART00000105545
|
arl3
|
ADP-ribosylation factor-like 3 |
| chr25_-_23052707 | 14.18 |
ENSDART00000024633
|
dusp8a
|
dual specificity phosphatase 8a |
| chr3_+_15271943 | 14.14 |
ENSDART00000141752
|
asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr25_+_16945348 | 14.13 |
ENSDART00000016591
|
fgf6a
|
fibroblast growth factor 6a |
| chr17_+_12698532 | 14.04 |
ENSDART00000064509
ENSDART00000136830 |
stmn4l
|
stathmin-like 4, like |
| chr20_-_37522569 | 13.95 |
ENSDART00000177011
ENSDART00000058502 |
hivep2a
|
human immunodeficiency virus type I enhancer binding protein 2a |
| chr7_-_40993456 | 13.89 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr2_+_20332044 | 13.86 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr9_+_42095220 | 13.72 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr7_+_19882066 | 13.63 |
ENSDART00000111144
|
TMEM151A
|
transmembrane protein 151A |
| chr17_-_35881841 | 13.63 |
ENSDART00000110040
|
sox11a
|
SRY (sex determining region Y)-box 11a |
| chr17_+_29345606 | 13.54 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr10_-_22249444 | 13.49 |
ENSDART00000148831
|
fgf11b
|
fibroblast growth factor 11b |
| chr3_+_33367954 | 13.43 |
ENSDART00000103161
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr23_-_28141419 | 13.36 |
ENSDART00000133039
|
tac3a
|
tachykinin 3a |
| chr7_-_48805181 | 13.24 |
ENSDART00000015884
|
mfge8a
|
milk fat globule-EGF factor 8 protein a |
| chr8_+_28066063 | 13.17 |
ENSDART00000078533
|
kcnd3
|
potassium voltage-gated channel, Shal-related subfamily, member 3 |
| chr10_+_15777064 | 13.14 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr20_-_18736281 | 13.13 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr15_+_28685892 | 12.78 |
ENSDART00000155815
ENSDART00000060244 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr13_-_33398735 | 12.78 |
ENSDART00000182601
ENSDART00000103628 |
btbd6a
|
BTB (POZ) domain containing 6a |
| chr2_-_27775236 | 12.71 |
ENSDART00000187983
|
XKR4
|
zgc:123035 |
| chr2_+_25929619 | 12.52 |
ENSDART00000137746
|
slc7a14a
|
solute carrier family 7, member 14a |
| chr1_+_54908895 | 12.51 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr23_+_19590598 | 12.44 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr14_-_7888748 | 12.42 |
ENSDART00000166293
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr19_-_10395683 | 12.38 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr13_+_15816573 | 12.37 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr2_+_20331445 | 12.34 |
ENSDART00000186880
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr2_-_11912347 | 12.26 |
ENSDART00000023851
|
abhd3
|
abhydrolase domain containing 3 |
| chr8_+_39634114 | 12.22 |
ENSDART00000144293
|
msi1
|
musashi RNA-binding protein 1 |
| chr4_+_26496489 | 12.20 |
ENSDART00000160652
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr25_-_31863374 | 12.19 |
ENSDART00000028338
|
scamp5a
|
secretory carrier membrane protein 5a |
| chr7_-_39203799 | 12.19 |
ENSDART00000173727
|
chrm4a
|
cholinergic receptor, muscarinic 4a |
| chr6_+_33537267 | 12.18 |
ENSDART00000040334
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr22_+_12366516 | 12.15 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr1_+_40023640 | 12.14 |
ENSDART00000101623
|
lgi2b
|
leucine-rich repeat LGI family, member 2b |
| chr7_-_71434298 | 12.10 |
ENSDART00000180507
|
lgi2a
|
leucine-rich repeat LGI family, member 2a |
| chr1_-_40227166 | 11.96 |
ENSDART00000146680
|
si:ch211-113e8.3
|
si:ch211-113e8.3 |
| chr5_-_23999777 | 11.82 |
ENSDART00000085969
|
map7d2a
|
MAP7 domain containing 2a |
| chr14_-_33454595 | 11.71 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr8_+_29267093 | 11.66 |
ENSDART00000077647
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr5_-_41560874 | 11.61 |
ENSDART00000136702
|
dnajb5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr25_-_31763897 | 11.47 |
ENSDART00000041740
|
ubl7a
|
ubiquitin-like 7a (bone marrow stromal cell-derived) |
| chr15_+_36115955 | 11.45 |
ENSDART00000032702
|
sst1.2
|
somatostatin 1, tandem duplicate 2 |
| chr11_-_29623380 | 11.44 |
ENSDART00000162587
ENSDART00000193935 ENSDART00000191646 |
chd5
|
chromodomain helicase DNA binding protein 5 |
| chr20_+_26095530 | 11.41 |
ENSDART00000139350
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr3_-_5964557 | 11.25 |
ENSDART00000184738
|
BX284638.1
|
|
| chr6_+_13933464 | 11.05 |
ENSDART00000109144
|
ptprnb
|
protein tyrosine phosphatase, receptor type, Nb |
| chr22_-_38621438 | 10.81 |
ENSDART00000098330
|
nppc
|
natriuretic peptide C |
| chr18_+_16749091 | 10.78 |
ENSDART00000061265
|
rnf141
|
ring finger protein 141 |
| chr7_-_51476276 | 10.75 |
ENSDART00000082464
|
nhsl2
|
NHS-like 2 |
| chr23_-_637347 | 10.74 |
ENSDART00000132175
|
l1camb
|
L1 cell adhesion molecule, paralog b |
| chr12_+_19305390 | 10.57 |
ENSDART00000183987
ENSDART00000066391 |
csnk1e
|
casein kinase 1, epsilon |
| chr18_-_21859019 | 10.57 |
ENSDART00000100885
|
nrn1la
|
neuritin 1-like a |
| chr20_+_19238382 | 10.56 |
ENSDART00000136757
|
fndc4a
|
fibronectin type III domain containing 4a |
| chr25_-_37084032 | 10.50 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr13_+_4505079 | 10.45 |
ENSDART00000144312
|
pde10a
|
phosphodiesterase 10A |
| chr23_+_28582865 | 10.39 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr7_+_15872357 | 10.36 |
ENSDART00000165757
|
pax6b
|
paired box 6b |
| chr11_-_17713987 | 10.31 |
ENSDART00000090401
|
fam19a4b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4b |
| chr14_-_1955257 | 10.14 |
ENSDART00000193254
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr14_-_2318590 | 10.04 |
ENSDART00000192735
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr3_+_40576447 | 10.02 |
ENSDART00000083212
|
fscn1a
|
fascin actin-bundling protein 1a |
| chr13_+_12174937 | 9.97 |
ENSDART00000171683
|
gabrg1
|
gamma-aminobutyric acid type A receptor gamma1 subunit |
| chr13_+_17702853 | 9.84 |
ENSDART00000145438
|
si:dkey-27m7.4
|
si:dkey-27m7.4 |
| chr18_-_22094102 | 9.69 |
ENSDART00000100904
|
pard6a
|
par-6 family cell polarity regulator alpha |
| chr5_-_46896541 | 9.69 |
ENSDART00000133240
|
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr9_-_31747106 | 9.67 |
ENSDART00000048469
ENSDART00000145204 ENSDART00000186889 |
nalcn
|
sodium leak channel, non-selective |
| chr9_-_18742704 | 9.64 |
ENSDART00000145401
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr20_-_39103119 | 9.64 |
ENSDART00000143379
|
rcan2
|
regulator of calcineurin 2 |
| chr2_+_21090317 | 9.61 |
ENSDART00000109568
ENSDART00000139633 |
pip4k2ab
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha b |
| chr14_-_1958994 | 9.60 |
ENSDART00000161783
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr9_+_21535885 | 9.52 |
ENSDART00000141408
|
arhgef7a
|
Rho guanine nucleotide exchange factor (GEF) 7a |
| chr16_+_4055331 | 9.51 |
ENSDART00000128978
|
CR391998.1
|
|
| chr11_+_15931188 | 9.48 |
ENSDART00000165768
ENSDART00000081098 |
draxin
|
dorsal inhibitory axon guidance protein |
| chr20_+_30490682 | 9.45 |
ENSDART00000184871
|
myt1la
|
myelin transcription factor 1-like, a |
| chr15_-_163586 | 9.45 |
ENSDART00000163597
|
SEPT4
|
septin-4 |
| chr10_-_7858553 | 9.41 |
ENSDART00000182010
|
inpp5ja
|
inositol polyphosphate-5-phosphatase Ja |
| chr14_-_2050057 | 9.38 |
ENSDART00000112875
|
PCDHB15
|
protocadherin beta 15 |
| chr12_+_7491690 | 9.36 |
ENSDART00000152564
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr19_+_45970692 | 9.19 |
ENSDART00000158781
|
si:ch211-153f2.7
|
si:ch211-153f2.7 |
| chr17_+_8184649 | 9.19 |
ENSDART00000091818
|
tulp4b
|
tubby like protein 4b |
| chr10_+_29698467 | 9.16 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr10_+_22782522 | 9.08 |
ENSDART00000079498
ENSDART00000145558 |
si:ch211-237l4.6
|
si:ch211-237l4.6 |
| chr25_-_8030113 | 9.05 |
ENSDART00000104674
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr1_+_51312752 | 8.99 |
ENSDART00000063938
|
mast1a
|
microtubule associated serine/threonine kinase 1a |
| chr3_+_32698424 | 8.98 |
ENSDART00000055340
|
fus
|
FUS RNA binding protein |
| chr17_+_18117029 | 8.92 |
ENSDART00000154646
ENSDART00000179739 |
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr13_+_24552254 | 8.89 |
ENSDART00000147907
|
lgalslb
|
lectin, galactoside-binding-like b |
| chr17_+_17955063 | 8.87 |
ENSDART00000104999
|
ccdc85ca
|
coiled-coil domain containing 85C, a |
| chr21_-_5856050 | 8.86 |
ENSDART00000115367
|
CABZ01071020.1
|
|
| chr17_+_24320861 | 8.81 |
ENSDART00000179858
|
otx1
|
orthodenticle homeobox 1 |
| chr5_-_20814576 | 8.77 |
ENSDART00000098682
ENSDART00000147639 |
si:ch211-225b11.1
|
si:ch211-225b11.1 |
| chr8_+_28065803 | 8.74 |
ENSDART00000178481
|
kcnd3
|
potassium voltage-gated channel, Shal-related subfamily, member 3 |
| chr9_-_18743012 | 8.71 |
ENSDART00000131626
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr2_-_5942115 | 8.67 |
ENSDART00000154489
|
tmem125b
|
transmembrane protein 125b |
| chr3_-_21137362 | 8.65 |
ENSDART00000104051
|
cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr4_-_20081621 | 8.65 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr18_+_17428506 | 8.58 |
ENSDART00000100223
|
zgc:91860
|
zgc:91860 |
| chr23_+_45584223 | 8.57 |
ENSDART00000149367
|
si:ch73-290k24.5
|
si:ch73-290k24.5 |
| chr6_+_3004972 | 8.49 |
ENSDART00000186750
ENSDART00000183862 ENSDART00000191485 ENSDART00000171014 |
ptprfa
|
protein tyrosine phosphatase, receptor type, f, a |
| chr24_-_22702017 | 8.47 |
ENSDART00000179403
|
ctnnd2a
|
catenin (cadherin-associated protein), delta 2a |
| chr18_-_18850302 | 8.36 |
ENSDART00000131965
ENSDART00000167624 |
tgm2l
|
transglutaminase 2, like |
| chr7_-_26532089 | 8.32 |
ENSDART00000121698
|
senp3b
|
SUMO1/sentrin/SMT3 specific peptidase 3b |
| chr2_+_9821757 | 8.31 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr13_+_28854438 | 8.26 |
ENSDART00000193407
ENSDART00000189554 |
CU639469.1
|
|
| chr20_+_522457 | 8.21 |
ENSDART00000138585
|
nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr22_+_18786797 | 8.19 |
ENSDART00000141864
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr24_-_35767501 | 8.16 |
ENSDART00000105680
ENSDART00000042290 ENSDART00000166264 |
dtna
|
dystrobrevin, alpha |
| chr20_-_46362606 | 7.88 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr10_+_5689510 | 7.85 |
ENSDART00000183217
ENSDART00000172632 |
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr12_-_10421955 | 7.82 |
ENSDART00000052004
|
zgc:153595
|
zgc:153595 |
| chr10_-_26163989 | 7.79 |
ENSDART00000136472
|
trim3b
|
tripartite motif containing 3b |
| chr6_+_52350443 | 7.67 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr10_-_26744131 | 7.64 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr22_-_16443199 | 7.58 |
ENSDART00000006290
ENSDART00000193335 |
plekhb2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr23_+_24611747 | 7.54 |
ENSDART00000134978
|
nckap5l
|
NCK-associated protein 5-like |
| chr2_-_16359042 | 7.54 |
ENSDART00000057216
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr9_+_35017702 | 7.49 |
ENSDART00000193640
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr18_-_20560007 | 7.31 |
ENSDART00000141367
ENSDART00000090186 |
si:ch211-238n5.4
|
si:ch211-238n5.4 |
| chr15_+_1372343 | 7.29 |
ENSDART00000152285
|
schip1
|
schwannomin interacting protein 1 |
| chr16_+_46459680 | 7.29 |
ENSDART00000101698
|
rpz3
|
rapunzel 3 |
| chr13_+_12175724 | 7.19 |
ENSDART00000166053
|
gabrg1
|
gamma-aminobutyric acid type A receptor gamma1 subunit |
| chr9_+_6997861 | 7.14 |
ENSDART00000190491
|
inpp4aa
|
inositol polyphosphate-4-phosphatase type I Aa |
| chr22_-_600016 | 7.09 |
ENSDART00000086434
|
tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr13_-_11035420 | 7.08 |
ENSDART00000108709
|
cep170aa
|
centrosomal protein 170Aa |
| chr12_+_33038757 | 7.08 |
ENSDART00000153146
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr1_-_54947592 | 7.06 |
ENSDART00000129710
|
crtac1a
|
cartilage acidic protein 1a |
| chr4_+_12111154 | 7.05 |
ENSDART00000036779
|
tmem178b
|
transmembrane protein 178B |
Network of associatons between targets according to the STRING database.
First level regulatory network of junba+junbb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 21.9 | GO:0071435 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 6.7 | 20.2 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 5.8 | 28.9 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 4.9 | 68.8 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 4.8 | 14.5 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 4.2 | 21.0 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 4.0 | 4.0 | GO:0072318 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 3.9 | 19.3 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 3.6 | 17.8 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 3.5 | 10.5 | GO:0046099 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 3.2 | 22.5 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 3.1 | 15.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 3.0 | 24.3 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 3.0 | 15.0 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 2.7 | 27.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 2.3 | 7.0 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 2.2 | 31.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 2.2 | 8.7 | GO:0071788 | endoplasmic reticulum tubular network maintenance(GO:0071788) |
| 2.1 | 6.2 | GO:0045887 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 2.1 | 14.5 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 2.0 | 10.0 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 1.8 | 84.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 1.7 | 6.9 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 1.7 | 11.7 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 1.6 | 4.8 | GO:1903673 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 1.6 | 39.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 1.6 | 6.2 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 1.5 | 6.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 1.5 | 21.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 1.5 | 10.4 | GO:0090104 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 1.5 | 21.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 1.4 | 36.2 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 1.4 | 20.0 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 1.4 | 39.2 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 1.3 | 7.8 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 1.3 | 43.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 1.3 | 9.1 | GO:1903306 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 1.3 | 3.8 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 1.2 | 28.6 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 1.2 | 19.8 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 1.2 | 13.6 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 1.2 | 7.3 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 1.2 | 4.8 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 1.2 | 9.4 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 1.1 | 19.4 | GO:0032418 | lysosome localization(GO:0032418) |
| 1.1 | 5.6 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 1.1 | 12.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 1.1 | 3.3 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 1.1 | 5.4 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 1.1 | 2.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 1.0 | 4.2 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 1.0 | 4.0 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 1.0 | 2.9 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 1.0 | 6.9 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 1.0 | 9.7 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 1.0 | 21.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 1.0 | 5.7 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 1.0 | 14.3 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.9 | 8.5 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.9 | 12.1 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.9 | 3.6 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.9 | 2.6 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.8 | 2.5 | GO:0061213 | regulation of neurotransmitter uptake(GO:0051580) regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.8 | 14.5 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.8 | 2.4 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.8 | 5.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.8 | 9.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.7 | 17.2 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.7 | 5.1 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.7 | 3.6 | GO:0072574 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.7 | 2.9 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.7 | 6.5 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) single-organism membrane invagination(GO:1902534) |
| 0.7 | 4.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.7 | 4.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.7 | 5.9 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.6 | 5.8 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.6 | 9.6 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.6 | 2.5 | GO:0018199 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.6 | 15.7 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.6 | 4.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.6 | 3.7 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.6 | 1.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.6 | 10.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.6 | 26.9 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.6 | 3.6 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.6 | 32.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.6 | 20.0 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.6 | 6.2 | GO:0072178 | pronephric duct morphogenesis(GO:0039023) nephric duct morphogenesis(GO:0072178) |
| 0.6 | 3.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.6 | 18.8 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.5 | 6.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.5 | 8.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.5 | 4.7 | GO:0072098 | anterior/posterior pattern specification involved in pronephros development(GO:0034672) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.5 | 3.6 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.5 | 14.6 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.5 | 3.5 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.5 | 7.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.5 | 7.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.5 | 6.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.5 | 2.8 | GO:1901739 | regulation of myoblast fusion(GO:1901739) |
| 0.5 | 17.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.5 | 14.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.5 | 8.8 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.4 | 4.5 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.4 | 3.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.4 | 15.9 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.4 | 2.6 | GO:1903826 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.4 | 4.3 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.4 | 4.2 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.4 | 5.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.4 | 5.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.4 | 29.6 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.4 | 3.2 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.4 | 8.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.4 | 1.1 | GO:0009838 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.4 | 4.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.4 | 3.7 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.4 | 1.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.4 | 1.4 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.3 | 35.1 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.3 | 1.7 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.3 | 31.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.3 | 19.0 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.3 | 0.9 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.3 | 2.2 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.3 | 2.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.3 | 7.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.3 | 8.3 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.3 | 2.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.3 | 1.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.3 | 5.4 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.3 | 7.1 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.3 | 1.8 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 2.3 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.3 | 5.2 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.3 | 8.4 | GO:0051085 | 'de novo' protein folding(GO:0006458) 'de novo' posttranslational protein folding(GO:0051084) chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.2 | 5.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.2 | 3.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.2 | 1.2 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.2 | 6.3 | GO:0007164 | establishment of planar polarity(GO:0001736) establishment of tissue polarity(GO:0007164) |
| 0.2 | 1.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 3.3 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.2 | 1.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 36.2 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.2 | 8.4 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.2 | 10.4 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.2 | 11.4 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.2 | 3.9 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.2 | 2.7 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 49.6 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.2 | 5.1 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.2 | 48.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 12.1 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.2 | 3.3 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.2 | 1.5 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.2 | 2.9 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.2 | 14.4 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.2 | 12.2 | GO:0001819 | positive regulation of cytokine production(GO:0001819) |
| 0.2 | 10.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.2 | 7.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 1.4 | GO:0050771 | negative regulation of axon extension(GO:0030517) negative regulation of axonogenesis(GO:0050771) |
| 0.2 | 0.5 | GO:0050879 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.2 | 4.6 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.2 | 1.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 3.6 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.1 | 10.0 | GO:0044344 | fibroblast growth factor receptor signaling pathway(GO:0008543) cellular response to fibroblast growth factor stimulus(GO:0044344) response to fibroblast growth factor(GO:0071774) |
| 0.1 | 2.4 | GO:0051897 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 3.6 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 1.8 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 18.4 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.1 | 4.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 4.1 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 1.7 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 0.8 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.7 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 2.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 22.0 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 3.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.9 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.1 | 3.0 | GO:0034250 | positive regulation of cellular amide metabolic process(GO:0034250) positive regulation of translation(GO:0045727) |
| 0.1 | 0.4 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 20.2 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.1 | 4.5 | GO:0016358 | dendrite development(GO:0016358) |
| 0.1 | 3.6 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.1 | 1.8 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.1 | 2.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 3.4 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.1 | 1.6 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.1 | 4.6 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 4.3 | GO:0006906 | vesicle fusion(GO:0006906) organelle membrane fusion(GO:0090174) |
| 0.1 | 2.7 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 | 3.2 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.1 | 8.8 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 0.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 1.5 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.1 | 10.6 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 0.6 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.1 | 1.9 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.1 | 1.2 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.1 | 4.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 4.1 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.1 | 0.9 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 7.8 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.1 | 21.5 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.1 | 1.5 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.1 | 2.5 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.1 | 3.2 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 2.2 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.0 | 0.7 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.3 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.2 | GO:0060547 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.0 | 5.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 2.2 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 0.9 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 3.5 | GO:0043065 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 0.9 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 4.2 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 2.2 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 0.1 | GO:0048640 | negative regulation of developmental growth(GO:0048640) |
| 0.0 | 0.8 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 2.7 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 12.1 | GO:0099536 | synaptic signaling(GO:0099536) |
| 0.0 | 1.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.5 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.6 | GO:0001757 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.0 | 0.3 | GO:0010138 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.0 | 0.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 4.1 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 2.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.5 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 1.0 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.3 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.3 | GO:0032355 | response to estradiol(GO:0032355) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.0 | 102.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 3.9 | 46.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 3.9 | 15.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 3.1 | 15.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 3.0 | 20.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 3.0 | 14.8 | GO:0005883 | neurofilament(GO:0005883) |
| 2.4 | 19.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 2.2 | 19.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 2.1 | 12.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 2.0 | 20.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 2.0 | 20.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 2.0 | 5.9 | GO:0033268 | node of Ranvier(GO:0033268) |
| 1.5 | 5.8 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 1.4 | 5.6 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 1.2 | 4.7 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 1.0 | 4.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.9 | 8.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.9 | 14.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.9 | 6.9 | GO:0034464 | BBSome(GO:0034464) |
| 0.9 | 112.4 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.8 | 15.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.8 | 2.3 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.8 | 28.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.7 | 11.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.7 | 6.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.7 | 3.3 | GO:0034657 | GID complex(GO:0034657) |
| 0.6 | 6.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.6 | 3.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.6 | 27.3 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.6 | 6.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.6 | 4.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.6 | 5.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.6 | 50.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.6 | 2.4 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.6 | 17.2 | GO:0032590 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.6 | 4.5 | GO:0035032 | extrinsic component of vacuolar membrane(GO:0000306) phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.5 | 17.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.5 | 1.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.5 | 2.4 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.5 | 3.7 | GO:0072487 | MSL complex(GO:0072487) |
| 0.5 | 1.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.5 | 2.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.4 | 3.6 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.4 | 5.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.4 | 3.6 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.4 | 1.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.4 | 2.7 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.4 | 4.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.4 | 8.3 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.4 | 2.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.3 | 3.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.3 | 41.3 | GO:0030425 | dendrite(GO:0030425) |
| 0.3 | 23.1 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.3 | 3.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.3 | 26.2 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.3 | 6.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.3 | 8.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 2.5 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.3 | 5.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 1.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 12.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 5.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.2 | 8.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 5.1 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 11.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.2 | 5.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 5.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 3.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 3.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 0.5 | GO:0098842 | postsynaptic early endosome(GO:0098842) postsynaptic endosome(GO:0098845) |
| 0.2 | 68.1 | GO:0043005 | neuron projection(GO:0043005) |
| 0.2 | 1.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 3.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 2.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 11.1 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 1.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.4 | GO:0030120 | vesicle coat(GO:0030120) |
| 0.1 | 0.8 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 8.3 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.1 | 6.0 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 4.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 6.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 2.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 3.3 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 8.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.5 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 25.2 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 2.5 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 1.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 7.3 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 1.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 0.4 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 1.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 4.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 11.0 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 2.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.3 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 3.0 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 1.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.0 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 16.4 | GO:0005829 | cytosol(GO:0005829) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.6 | GO:0031834 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 4.1 | 12.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 3.9 | 15.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 3.7 | 21.9 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 3.5 | 10.5 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 3.0 | 15.0 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 2.8 | 19.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 2.5 | 32.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 2.4 | 21.9 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 2.4 | 14.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 2.4 | 95.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 2.3 | 11.7 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 2.2 | 31.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 2.1 | 10.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 2.1 | 12.4 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 2.0 | 8.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 2.0 | 10.2 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 2.0 | 14.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 2.0 | 7.8 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 1.6 | 14.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 1.5 | 23.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 1.5 | 41.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 1.5 | 11.9 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 1.4 | 17.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.3 | 20.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 1.2 | 17.2 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 1.2 | 40.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 1.2 | 23.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 1.2 | 9.4 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 1.2 | 28.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 1.1 | 29.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 1.1 | 5.6 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 1.1 | 12.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 1.1 | 5.4 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 1.1 | 25.7 | GO:0031267 | small GTPase binding(GO:0031267) |
| 1.0 | 4.0 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 1.0 | 9.7 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 1.0 | 13.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.9 | 4.6 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.8 | 8.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.8 | 14.3 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.8 | 3.0 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.7 | 11.9 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.7 | 6.0 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.7 | 4.6 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.6 | 25.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.6 | 2.5 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.6 | 3.6 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.6 | 34.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.6 | 3.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.6 | 14.5 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.5 | 9.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.5 | 8.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.5 | 26.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.5 | 33.2 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.5 | 2.5 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.5 | 3.9 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.5 | 2.4 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.5 | 19.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.5 | 4.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.4 | 4.8 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.4 | 4.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.4 | 5.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.4 | 15.5 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.4 | 8.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.4 | 42.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.4 | 6.9 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.4 | 8.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.4 | 1.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.4 | 11.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.4 | 2.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.4 | 12.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.4 | 17.2 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.4 | 10.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.4 | 1.4 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.3 | 4.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.3 | 4.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.3 | 4.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 20.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.3 | 3.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 14.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.3 | 2.4 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.3 | 8.4 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.3 | 5.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.3 | 10.9 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.3 | 25.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.3 | 5.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.3 | 6.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 16.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.3 | 4.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 3.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 5.7 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.2 | 0.9 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.2 | 0.7 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.2 | 38.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.2 | 3.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 4.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 5.1 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.2 | 8.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 5.0 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.2 | 6.2 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.2 | 0.6 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 9.1 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.2 | 3.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 14.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.2 | 3.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.2 | 2.5 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.2 | 0.7 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.2 | 9.7 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.2 | 13.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 56.3 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 15.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 1.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 11.3 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 1.9 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 3.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 2.0 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 7.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 2.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 3.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 5.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 1.5 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 23.5 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 1.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 2.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 0.5 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 0.3 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADPH binding(GO:0070402) NADH binding(GO:0070404) |
| 0.1 | 11.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 7.8 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 1.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 2.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 6.1 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 1.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 4.6 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 5.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 1.6 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 15.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) cis-trans isomerase activity(GO:0016859) |
| 0.0 | 20.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 2.0 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 1.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 2.4 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.6 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 10.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 20.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 1.1 | 23.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.8 | 17.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.7 | 14.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.7 | 15.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.6 | 6.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.5 | 8.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.5 | 15.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.4 | 9.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.3 | 3.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 5.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.3 | 4.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.3 | 1.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 12.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 4.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 3.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 5.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 1.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 4.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 6.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 6.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 3.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 3.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 5.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 2.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 3.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 3.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.8 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 2.2 | 36.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 1.1 | 14.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.9 | 15.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.7 | 34.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.7 | 10.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.7 | 5.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.6 | 5.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.5 | 3.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.5 | 4.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.5 | 39.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.5 | 16.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.5 | 4.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.4 | 3.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.4 | 5.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.4 | 5.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.4 | 5.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.4 | 14.1 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.3 | 3.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 3.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 4.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 4.1 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 7.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.2 | 9.7 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.2 | 2.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 1.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 4.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.2 | 5.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 3.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 2.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 3.1 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 2.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 2.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 9.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 0.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.4 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 3.5 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 5.2 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 1.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 4.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 2.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 3.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 5.1 | REACTOME NEURONAL SYSTEM | Genes involved in Neuronal System |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 1.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |