Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
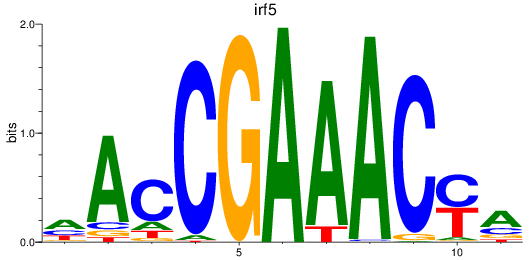
Results for irf5
Z-value: 3.56
Transcription factors associated with irf5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irf5
|
ENSDARG00000045681 | interferon regulatory factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irf5 | dr11_v1_chr4_-_13613148_13613148 | 0.80 | 8.6e-22 | Click! |
Activity profile of irf5 motif
Sorted Z-values of irf5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_2039511 | 90.47 |
ENSDART00000160223
|
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr10_-_22150419 | 57.52 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr15_-_2640966 | 56.23 |
ENSDART00000063320
|
cldne
|
claudin e |
| chr17_-_22552678 | 53.50 |
ENSDART00000079401
|
si:ch211-125o16.4
|
si:ch211-125o16.4 |
| chr14_-_33277743 | 52.94 |
ENSDART00000048130
|
stard14
|
START domain containing 14 |
| chr9_-_30264415 | 50.37 |
ENSDART00000060150
|
mid1ip1a
|
MID1 interacting protein 1a |
| chr8_+_44475793 | 44.04 |
ENSDART00000190118
|
si:ch73-211l2.3
|
si:ch73-211l2.3 |
| chr14_-_33278084 | 43.14 |
ENSDART00000132850
|
stard14
|
START domain containing 14 |
| chr7_+_69187585 | 42.01 |
ENSDART00000160499
ENSDART00000166258 |
marveld3
|
MARVEL domain containing 3 |
| chr15_-_33964897 | 37.84 |
ENSDART00000172075
ENSDART00000158126 ENSDART00000160456 |
lsr
|
lipolysis stimulated lipoprotein receptor |
| chr23_-_29556844 | 37.71 |
ENSDART00000138021
|
rbp7a
|
retinol binding protein 7a, cellular |
| chr17_+_30894431 | 37.29 |
ENSDART00000127996
|
degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr18_+_44532370 | 36.97 |
ENSDART00000086952
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr24_-_26310854 | 34.21 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr7_+_19483277 | 33.80 |
ENSDART00000173750
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr5_-_29531948 | 33.51 |
ENSDART00000098360
|
arrdc1a
|
arrestin domain containing 1a |
| chr7_+_49681040 | 30.73 |
ENSDART00000176372
ENSDART00000192172 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr10_+_15064433 | 30.08 |
ENSDART00000179978
|
parm1
|
prostate androgen-regulated mucin-like protein 1 |
| chr19_-_15420678 | 29.69 |
ENSDART00000151454
ENSDART00000027697 |
serinc2
|
serine incorporator 2 |
| chr22_-_17688868 | 29.28 |
ENSDART00000012336
ENSDART00000147070 |
tjp3
|
tight junction protein 3 |
| chr20_+_26683933 | 29.28 |
ENSDART00000139852
ENSDART00000077751 |
foxq1b
|
forkhead box Q1b |
| chr16_+_19637384 | 29.19 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr12_+_23424108 | 28.32 |
ENSDART00000077732
|
bambia
|
BMP and activin membrane-bound inhibitor (Xenopus laevis) homolog a |
| chr20_-_26551210 | 26.82 |
ENSDART00000077715
|
si:dkey-25e12.3
|
si:dkey-25e12.3 |
| chr2_-_21438492 | 26.37 |
ENSDART00000046098
|
plcd1b
|
phospholipase C, delta 1b |
| chr21_-_25295087 | 26.10 |
ENSDART00000087910
ENSDART00000147860 |
st14b
|
suppression of tumorigenicity 14 (colon carcinoma) b |
| chr7_+_19482877 | 25.97 |
ENSDART00000077868
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr4_-_11580948 | 25.71 |
ENSDART00000049066
|
net1
|
neuroepithelial cell transforming 1 |
| chr4_-_17669881 | 25.46 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr3_+_21200763 | 25.39 |
ENSDART00000067841
|
zgc:112038
|
zgc:112038 |
| chr20_+_6535176 | 25.10 |
ENSDART00000054652
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr16_-_41990421 | 24.21 |
ENSDART00000055921
|
pycard
|
PYD and CARD domain containing |
| chr1_+_58332000 | 23.93 |
ENSDART00000145234
|
ggt1l2.1
|
gamma-glutamyltransferase 1 like 2.1 |
| chr18_+_44532668 | 23.68 |
ENSDART00000140672
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr18_+_44532199 | 23.48 |
ENSDART00000135386
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr6_+_49771626 | 23.38 |
ENSDART00000134207
|
ctsz
|
cathepsin Z |
| chr14_+_33329420 | 23.03 |
ENSDART00000171090
ENSDART00000164062 |
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr6_+_49771372 | 22.99 |
ENSDART00000063251
|
ctsz
|
cathepsin Z |
| chr18_+_44532883 | 22.50 |
ENSDART00000121994
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr16_+_29492937 | 22.15 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr23_-_31266586 | 21.22 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr20_+_6535438 | 20.81 |
ENSDART00000145763
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr12_+_17154655 | 20.73 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr21_-_22543611 | 20.73 |
ENSDART00000177084
|
myo5b
|
myosin VB |
| chr24_-_34680956 | 20.01 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr23_+_2666944 | 19.90 |
ENSDART00000192861
|
CABZ01057928.1
|
|
| chr7_-_26579465 | 19.61 |
ENSDART00000173820
|
si:dkey-62k3.6
|
si:dkey-62k3.6 |
| chr2_-_37140423 | 19.45 |
ENSDART00000144220
|
tspan37
|
tetraspanin 37 |
| chr5_-_37959874 | 19.27 |
ENSDART00000031719
|
mpzl2b
|
myelin protein zero-like 2b |
| chr5_+_66326004 | 19.13 |
ENSDART00000144351
|
malt1
|
MALT paracaspase 1 |
| chr11_-_7261717 | 19.07 |
ENSDART00000128959
|
zgc:113223
|
zgc:113223 |
| chr5_-_12587053 | 18.97 |
ENSDART00000162780
|
vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr1_+_38153944 | 18.66 |
ENSDART00000135666
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr5_-_26187097 | 18.12 |
ENSDART00000137027
|
ccdc125
|
coiled-coil domain containing 125 |
| chr10_-_15879569 | 18.07 |
ENSDART00000136789
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr3_-_40976288 | 17.93 |
ENSDART00000193553
|
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr14_+_22076596 | 17.89 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr1_-_7570181 | 17.60 |
ENSDART00000103588
|
mxa
|
myxovirus (influenza) resistance A |
| chr10_+_5060191 | 17.45 |
ENSDART00000145908
ENSDART00000122397 |
carm1l
|
coactivator-associated arginine methyltransferase 1, like |
| chr16_-_45152169 | 17.24 |
ENSDART00000171452
|
si:ch73-343g19.4
|
si:ch73-343g19.4 |
| chr14_+_16345003 | 17.07 |
ENSDART00000003040
ENSDART00000165193 |
itln3
|
intelectin 3 |
| chr9_+_3055566 | 17.05 |
ENSDART00000189906
ENSDART00000175891 ENSDART00000093021 |
ppp1r9ala
|
protein phosphatase 1 regulatory subunit 9A-like A |
| chr7_-_20103384 | 15.96 |
ENSDART00000052902
|
acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr19_-_7690975 | 15.50 |
ENSDART00000151384
|
si:dkey-204a24.10
|
si:dkey-204a24.10 |
| chr18_+_45666489 | 15.09 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr3_-_40976463 | 14.98 |
ENSDART00000128450
ENSDART00000018676 |
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr3_-_29941357 | 14.98 |
ENSDART00000147732
ENSDART00000137973 ENSDART00000103523 |
grna
|
granulin a |
| chr9_+_23765587 | 14.89 |
ENSDART00000145120
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr9_+_41156818 | 14.81 |
ENSDART00000105764
ENSDART00000147052 |
stat4
|
signal transducer and activator of transcription 4 |
| chr22_+_19218733 | 14.59 |
ENSDART00000183212
ENSDART00000133595 |
si:dkey-21e2.7
|
si:dkey-21e2.7 |
| chr14_+_33329761 | 14.20 |
ENSDART00000161138
|
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr11_+_37251825 | 14.18 |
ENSDART00000169804
|
il17rc
|
interleukin 17 receptor C |
| chr19_+_14113886 | 14.08 |
ENSDART00000169343
|
kdf1b
|
keratinocyte differentiation factor 1b |
| chr3_+_32125452 | 13.96 |
ENSDART00000110396
|
zgc:194125
|
zgc:194125 |
| chr7_-_67214972 | 13.87 |
ENSDART00000156861
|
swap70a
|
switching B cell complex subunit SWAP70a |
| chr14_+_32918484 | 13.82 |
ENSDART00000105721
|
lnx2b
|
ligand of numb-protein X 2b |
| chr16_+_35595312 | 13.81 |
ENSDART00000170438
|
si:ch211-1i11.3
|
si:ch211-1i11.3 |
| chr20_+_36806398 | 13.70 |
ENSDART00000153317
|
abracl
|
ABRA C-terminal like |
| chr3_+_25191467 | 13.68 |
ENSDART00000156956
ENSDART00000154799 |
il2rb
|
interleukin 2 receptor, beta |
| chr17_-_30666037 | 13.66 |
ENSDART00000156509
|
alkal2b
|
ALK and LTK ligand 2b |
| chr3_+_32571929 | 13.65 |
ENSDART00000151025
|
si:ch73-248e21.1
|
si:ch73-248e21.1 |
| chr15_-_29162193 | 13.64 |
ENSDART00000138449
ENSDART00000099885 |
xaf1
|
XIAP associated factor 1 |
| chr7_-_19614916 | 13.58 |
ENSDART00000169029
|
zgc:194655
|
zgc:194655 |
| chr23_+_42346799 | 13.30 |
ENSDART00000159985
ENSDART00000172144 |
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr23_+_642001 | 13.22 |
ENSDART00000030643
ENSDART00000124850 |
irf10
|
interferon regulatory factor 10 |
| chr3_+_29941777 | 13.21 |
ENSDART00000113889
|
ifi35
|
interferon-induced protein 35 |
| chr4_+_12931763 | 13.18 |
ENSDART00000016382
|
wif1
|
wnt inhibitory factor 1 |
| chr5_+_50913357 | 12.89 |
ENSDART00000092938
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr9_-_39968820 | 12.86 |
ENSDART00000100311
|
IKZF2
|
si:zfos-1425h8.1 |
| chr10_+_4046448 | 12.77 |
ENSDART00000123086
ENSDART00000052268 |
pitpnb
|
phosphatidylinositol transfer protein, beta |
| chr23_+_642395 | 12.53 |
ENSDART00000186995
|
irf10
|
interferon regulatory factor 10 |
| chr16_+_42772678 | 12.40 |
ENSDART00000155575
|
si:ch211-135n15.2
|
si:ch211-135n15.2 |
| chr1_+_58290933 | 12.24 |
ENSDART00000140536
|
si:dkey-222h21.6
|
si:dkey-222h21.6 |
| chr15_-_40175894 | 12.09 |
ENSDART00000156632
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr22_-_30973791 | 11.99 |
ENSDART00000104728
|
ssuh2.2
|
ssu-2 homolog, tandem duplicate 2 |
| chr14_-_1432875 | 11.90 |
ENSDART00000164408
ENSDART00000159774 |
zgc:152774
|
zgc:152774 |
| chr4_-_77557279 | 11.73 |
ENSDART00000180113
|
AL935186.10
|
|
| chr3_+_32139666 | 11.70 |
ENSDART00000109791
|
zgc:92066
|
zgc:92066 |
| chr10_+_35358675 | 11.69 |
ENSDART00000193263
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr10_-_2971407 | 11.56 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr1_-_29747702 | 11.33 |
ENSDART00000133225
ENSDART00000189670 |
spp2
|
secreted phosphoprotein 2 |
| chr25_+_19720228 | 11.27 |
ENSDART00000181804
|
mxd
|
myxovirus (influenza virus) resistance D |
| chr2_-_41861040 | 11.21 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
| chr25_+_16646113 | 11.19 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr2_-_24554416 | 11.18 |
ENSDART00000052061
|
cnn2
|
calponin 2 |
| chr12_-_23658888 | 11.13 |
ENSDART00000088319
|
map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr5_+_58455488 | 11.09 |
ENSDART00000038602
ENSDART00000127958 |
slc37a2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr14_-_28001986 | 11.09 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr25_+_28862660 | 10.87 |
ENSDART00000154681
|
si:ch211-106e7.2
|
si:ch211-106e7.2 |
| chr16_+_46497149 | 10.80 |
ENSDART00000135151
ENSDART00000058324 |
rpz4
|
rapunzel 4 |
| chr6_+_48348415 | 10.79 |
ENSDART00000064826
|
mov10a
|
Mov10 RISC complex RNA helicase a |
| chr3_+_25999477 | 10.62 |
ENSDART00000024316
|
mcm5
|
minichromosome maintenance complex component 5 |
| chr23_-_30045661 | 10.61 |
ENSDART00000122239
ENSDART00000103480 |
ccdc187
|
coiled-coil domain containing 187 |
| chr5_-_30382925 | 10.51 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr15_+_11883804 | 10.50 |
ENSDART00000163255
|
gpr184
|
G protein-coupled receptor 184 |
| chr22_-_11648094 | 10.49 |
ENSDART00000191791
|
dpp4
|
dipeptidyl-peptidase 4 |
| chr12_-_33805366 | 10.31 |
ENSDART00000030566
|
galk1
|
galactokinase 1 |
| chr10_+_2742499 | 10.17 |
ENSDART00000122847
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr14_-_33585809 | 10.12 |
ENSDART00000023540
|
sash3
|
SAM and SH3 domain containing 3 |
| chr25_-_12809361 | 10.11 |
ENSDART00000162750
|
ca5a
|
carbonic anhydrase Va |
| chr8_-_27687095 | 10.05 |
ENSDART00000086946
|
mov10b.1
|
Moloney leukemia virus 10b, tandem duplicate 1 |
| chr5_-_69716501 | 9.80 |
ENSDART00000158956
|
mob1a
|
MOB kinase activator 1A |
| chr7_+_13995792 | 9.61 |
ENSDART00000091470
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr13_+_30692669 | 9.58 |
ENSDART00000187818
|
CR762483.1
|
|
| chr2_+_47927026 | 9.47 |
ENSDART00000143023
|
ftr25
|
finTRIM family, member 25 |
| chr11_-_42472941 | 9.47 |
ENSDART00000166624
|
arf4b
|
ADP-ribosylation factor 4b |
| chr24_+_11334733 | 9.47 |
ENSDART00000147552
ENSDART00000143171 |
si:dkey-12l12.1
|
si:dkey-12l12.1 |
| chr8_+_45361775 | 9.41 |
ENSDART00000015193
|
chmp4bb
|
charged multivesicular body protein 4Bb |
| chr6_-_1777831 | 9.31 |
ENSDART00000167390
|
zgc:113442
|
zgc:113442 |
| chr4_+_8569199 | 9.30 |
ENSDART00000165181
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr9_+_23003208 | 9.13 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr4_-_68563862 | 8.92 |
ENSDART00000182970
|
BX548011.2
|
|
| chr4_-_20222182 | 8.86 |
ENSDART00000132464
|
gsap
|
gamma-secretase activating protein |
| chr11_+_10909183 | 8.65 |
ENSDART00000064860
|
rbms1a
|
RNA binding motif, single stranded interacting protein 1a |
| chr7_-_24149670 | 8.62 |
ENSDART00000005884
|
mmp14a
|
matrix metallopeptidase 14a (membrane-inserted) |
| chr23_-_29553430 | 8.44 |
ENSDART00000157773
ENSDART00000126384 |
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr2_-_55797318 | 8.41 |
ENSDART00000158147
|
calr3b
|
calreticulin 3b |
| chr18_+_27337994 | 8.38 |
ENSDART00000136172
|
si:dkey-29p10.4
|
si:dkey-29p10.4 |
| chr22_-_17474583 | 8.27 |
ENSDART00000148027
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr16_+_33144306 | 8.25 |
ENSDART00000101953
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr15_+_25452092 | 8.21 |
ENSDART00000009545
|
pak4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr10_+_2587234 | 8.09 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr17_+_1627379 | 8.09 |
ENSDART00000184050
|
LO018432.1
|
|
| chr2_-_47870649 | 8.05 |
ENSDART00000142854
|
ftr05
|
finTRIM family, member 5 |
| chr9_-_23765480 | 8.05 |
ENSDART00000027212
|
si:ch211-219a4.6
|
si:ch211-219a4.6 |
| chr7_+_19615056 | 8.05 |
ENSDART00000124752
ENSDART00000190297 |
si:ch211-212k18.15
|
si:ch211-212k18.15 |
| chr21_+_43253538 | 7.97 |
ENSDART00000179940
ENSDART00000164806 ENSDART00000147026 |
shroom1
|
shroom family member 1 |
| chr5_+_21922534 | 7.76 |
ENSDART00000148092
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr13_+_24263049 | 7.72 |
ENSDART00000135992
ENSDART00000088005 |
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr11_-_13107106 | 7.71 |
ENSDART00000184477
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr24_+_22731228 | 7.70 |
ENSDART00000146733
|
si:dkey-225k4.1
|
si:dkey-225k4.1 |
| chr3_+_32135037 | 7.59 |
ENSDART00000110490
|
BX511021.1
|
|
| chr22_-_30678518 | 7.51 |
ENSDART00000138282
|
si:dkey-42l16.1
|
si:dkey-42l16.1 |
| chr22_-_17474781 | 7.51 |
ENSDART00000186817
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr3_+_36127287 | 7.42 |
ENSDART00000058605
ENSDART00000182500 |
scpep1
|
serine carboxypeptidase 1 |
| chr18_+_17624295 | 7.33 |
ENSDART00000151865
ENSDART00000190408 ENSDART00000189204 ENSDART00000188894 ENSDART00000033762 ENSDART00000193471 |
nlrc5
|
NLR family, CARD domain containing 5 |
| chr18_+_45708744 | 7.21 |
ENSDART00000077341
|
depdc7
|
DEP domain containing 7 |
| chr12_-_27242498 | 7.11 |
ENSDART00000152609
ENSDART00000152170 |
si:dkey-11c5.11
|
si:dkey-11c5.11 |
| chr2_-_13333932 | 7.06 |
ENSDART00000150238
ENSDART00000168258 |
si:dkey-185p13.1
vps4b
|
si:dkey-185p13.1 vacuolar protein sorting 4b homolog B (S. cerevisiae) |
| chr16_-_30903930 | 7.05 |
ENSDART00000143996
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr8_+_29986265 | 7.01 |
ENSDART00000148258
|
ptch1
|
patched 1 |
| chr18_+_40467719 | 7.00 |
ENSDART00000087647
|
ugt5c2
|
UDP glucuronosyltransferase 5 family, polypeptide C2 |
| chr4_+_76509294 | 6.95 |
ENSDART00000099899
|
ms4a17a.17
|
membrane-spanning 4-domains, subfamily A, member 17A.17 |
| chr24_-_26885897 | 6.87 |
ENSDART00000180512
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr13_+_15182149 | 6.81 |
ENSDART00000193644
ENSDART00000134421 ENSDART00000086281 |
mavs
|
mitochondrial antiviral signaling protein |
| chr3_-_29508959 | 6.76 |
ENSDART00000055408
|
cyth4a
|
cytohesin 4a |
| chr15_-_33818872 | 6.75 |
ENSDART00000158325
|
n4bp2l2
|
NEDD4 binding protein 2-like 2 |
| chr15_-_31067589 | 6.73 |
ENSDART00000060157
|
lgals9l3
|
lectin, galactoside-binding, soluble, 9 (galectin 9)-like 3 |
| chr6_-_18400548 | 6.59 |
ENSDART00000179797
ENSDART00000164891 |
trim25
|
tripartite motif containing 25 |
| chr12_-_41759686 | 6.49 |
ENSDART00000172175
ENSDART00000165152 |
ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta |
| chr6_-_35401282 | 6.43 |
ENSDART00000127612
|
rgs5a
|
regulator of G protein signaling 5a |
| chr5_+_35561607 | 6.36 |
ENSDART00000098022
|
ar
|
androgen receptor |
| chr22_+_38049130 | 6.36 |
ENSDART00000097533
|
wwtr1
|
WW domain containing transcription regulator 1 |
| chr15_-_17071328 | 6.26 |
ENSDART00000122617
|
si:ch211-24o10.6
|
si:ch211-24o10.6 |
| chr25_+_20272145 | 6.23 |
ENSDART00000109605
|
si:dkey-219c3.2
|
si:dkey-219c3.2 |
| chr18_-_16922905 | 6.21 |
ENSDART00000187165
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr24_+_37688729 | 6.14 |
ENSDART00000137017
|
h3f3d
|
H3 histone, family 3D |
| chr16_-_25829779 | 6.06 |
ENSDART00000086301
|
irge4
|
immunity-related GTPase family, e4 |
| chr5_+_57442271 | 6.06 |
ENSDART00000097395
|
prpf4
|
PRP4 pre-mRNA processing factor 4 homolog (yeast) |
| chr25_+_5983430 | 6.06 |
ENSDART00000074814
|
ppib
|
peptidylprolyl isomerase B (cyclophilin B) |
| chr23_-_45501177 | 6.02 |
ENSDART00000150103
|
col24a1
|
collagen type XXIV alpha 1 |
| chr3_+_24189804 | 5.99 |
ENSDART00000134723
|
prr15la
|
proline rich 15-like a |
| chr2_-_6115688 | 5.98 |
ENSDART00000081663
|
prdx1
|
peroxiredoxin 1 |
| chr6_-_33916756 | 5.95 |
ENSDART00000137447
ENSDART00000138488 |
nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr24_+_7782313 | 5.90 |
ENSDART00000111090
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr22_+_24623936 | 5.87 |
ENSDART00000160924
|
mcoln2
|
mucolipin 2 |
| chr12_+_27243059 | 5.80 |
ENSDART00000066269
|
arl4d
|
ADP-ribosylation factor-like 4D |
| chr15_-_21702317 | 5.74 |
ENSDART00000155824
|
IL4I1
|
si:dkey-40g16.6 |
| chr11_+_42478184 | 5.69 |
ENSDART00000089963
|
zgc:110286
|
zgc:110286 |
| chr12_-_35936329 | 5.61 |
ENSDART00000166634
|
rnf213b
|
ring finger protein 213b |
| chr3_-_55650771 | 5.60 |
ENSDART00000162413
|
axin2
|
axin 2 (conductin, axil) |
| chr23_+_44349252 | 5.38 |
ENSDART00000097644
|
ephb4b
|
eph receptor B4b |
| chr25_-_774350 | 5.36 |
ENSDART00000166321
ENSDART00000160386 |
IRAK4
|
interleukin 1 receptor associated kinase 4 |
| chr6_-_49983329 | 5.29 |
ENSDART00000113083
|
asip1
|
agouti signaling protein 1 |
| chr17_+_34805897 | 5.28 |
ENSDART00000137090
ENSDART00000077626 |
id2a
|
inhibitor of DNA binding 2a |
| chr3_+_3046909 | 5.28 |
ENSDART00000122435
|
BX004816.2
|
|
| chr13_+_22863516 | 5.21 |
ENSDART00000113082
ENSDART00000189200 |
hnrnph3
|
heterogeneous nuclear ribonucleoprotein H3 (2H9) |
| chr18_+_37015185 | 5.21 |
ENSDART00000191305
|
sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr21_+_19844195 | 5.17 |
ENSDART00000065674
ENSDART00000157188 |
fybb
|
FYN binding protein b |
| chr22_+_336256 | 5.17 |
ENSDART00000019155
|
btg2
|
B-cell translocation gene 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of irf5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.0 | 42.0 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 10.0 | 30.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 9.6 | 57.5 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 5.2 | 20.7 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 3.7 | 11.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 3.1 | 9.4 | GO:0044878 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 3.0 | 23.9 | GO:0031179 | peptide modification(GO:0031179) |
| 2.9 | 29.3 | GO:0030104 | water homeostasis(GO:0030104) |
| 2.8 | 14.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 2.7 | 13.4 | GO:0039528 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 2.7 | 37.3 | GO:0034311 | diol metabolic process(GO:0034311) |
| 2.3 | 90.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 2.3 | 13.7 | GO:0070376 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 2.2 | 17.6 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 2.0 | 10.0 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 2.0 | 17.9 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 1.9 | 15.0 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 1.8 | 5.3 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 1.5 | 6.0 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 1.5 | 10.3 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 1.4 | 25.7 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 1.4 | 18.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 1.3 | 6.7 | GO:0035739 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 1.3 | 5.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 1.3 | 6.4 | GO:0007618 | mating(GO:0007618) male gonad development(GO:0008584) |
| 1.3 | 6.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 1.2 | 47.3 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 1.2 | 56.2 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 1.2 | 7.3 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 1.2 | 4.8 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 1.1 | 5.6 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 1.1 | 11.2 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 1.1 | 28.3 | GO:0030168 | platelet activation(GO:0030168) |
| 1.1 | 34.2 | GO:0007568 | aging(GO:0007568) |
| 1.0 | 3.1 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 1.0 | 17.4 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 1.0 | 9.1 | GO:0014812 | muscle cell migration(GO:0014812) |
| 1.0 | 6.0 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 1.0 | 8.9 | GO:0042987 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 1.0 | 10.6 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.9 | 13.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.9 | 18.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.9 | 7.0 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.8 | 141.6 | GO:0051604 | protein maturation(GO:0051604) |
| 0.8 | 4.8 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.8 | 5.3 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.8 | 42.9 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.7 | 4.9 | GO:0014028 | notochord formation(GO:0014028) |
| 0.7 | 13.6 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.7 | 11.2 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.6 | 6.5 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.6 | 19.5 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.6 | 1.8 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.6 | 4.8 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.6 | 6.0 | GO:0019430 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.6 | 19.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.5 | 4.3 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.5 | 20.9 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.5 | 8.8 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.5 | 7.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.5 | 17.3 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.5 | 37.8 | GO:0007492 | endoderm development(GO:0007492) |
| 0.5 | 3.4 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.5 | 1.9 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.5 | 7.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.4 | 4.0 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.4 | 13.7 | GO:2001236 | regulation of extrinsic apoptotic signaling pathway(GO:2001236) |
| 0.4 | 12.8 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.4 | 2.0 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.4 | 2.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.4 | 13.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.4 | 1.2 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.4 | 11.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.4 | 4.9 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.4 | 7.1 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.4 | 3.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.4 | 22.1 | GO:0060348 | bone development(GO:0060348) |
| 0.3 | 2.8 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.3 | 20.0 | GO:0001894 | tissue homeostasis(GO:0001894) |
| 0.3 | 2.7 | GO:0061316 | canonical Wnt signaling pathway involved in heart development(GO:0061316) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.3 | 2.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 4.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.3 | 16.0 | GO:0007259 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.3 | 2.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.3 | 26.4 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.3 | 4.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.3 | 4.7 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.3 | 3.5 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.3 | 1.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.3 | 13.3 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.3 | 1.5 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.2 | 3.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 10.5 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.2 | 39.6 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.2 | 10.8 | GO:0035194 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.2 | 5.0 | GO:0048920 | neuromast primordium migration(GO:0048883) posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.2 | 21.7 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.2 | 8.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.2 | 11.2 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.2 | 6.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 21.1 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.2 | 18.3 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.2 | 27.9 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.2 | 12.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.2 | 4.8 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.2 | 4.8 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.2 | 29.2 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.2 | 34.6 | GO:0072594 | establishment of protein localization to organelle(GO:0072594) |
| 0.2 | 3.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 | 8.1 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.2 | 6.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 6.2 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 8.2 | GO:0002573 | myeloid leukocyte differentiation(GO:0002573) |
| 0.1 | 11.6 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 3.6 | GO:0006458 | 'de novo' protein folding(GO:0006458) 'de novo' posttranslational protein folding(GO:0051084) chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 1.4 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 2.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 4.5 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.1 | 3.8 | GO:0030301 | cholesterol transport(GO:0030301) |
| 0.1 | 39.2 | GO:0001525 | angiogenesis(GO:0001525) |
| 0.1 | 11.4 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.1 | 3.1 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.1 | 1.4 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 5.2 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 6.7 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.1 | 3.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 1.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 7.7 | GO:0030218 | erythrocyte differentiation(GO:0030218) erythrocyte homeostasis(GO:0034101) |
| 0.1 | 3.2 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 2.4 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 4.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 61.4 | GO:0006508 | proteolysis(GO:0006508) |
| 0.1 | 1.0 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 15.9 | GO:0032970 | regulation of actin filament-based process(GO:0032970) |
| 0.0 | 0.4 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 1.8 | GO:0045786 | negative regulation of cell cycle(GO:0045786) |
| 0.0 | 0.8 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 5.9 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 6.5 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 3.5 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 53.5 | GO:0043034 | costamere(GO:0043034) |
| 6.1 | 24.2 | GO:0061702 | inflammasome complex(GO:0061702) |
| 2.9 | 11.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 2.0 | 6.0 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 1.7 | 10.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 1.7 | 203.1 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 1.6 | 11.2 | GO:0016234 | inclusion body(GO:0016234) |
| 1.4 | 26.4 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 1.1 | 9.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 1.1 | 20.0 | GO:0030057 | desmosome(GO:0030057) |
| 1.0 | 74.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 1.0 | 4.8 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.9 | 11.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.8 | 3.1 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.7 | 4.8 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.7 | 2.0 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.6 | 11.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.6 | 20.8 | GO:0043186 | P granule(GO:0043186) |
| 0.5 | 37.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.5 | 8.0 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.5 | 1.4 | GO:1990879 | CST complex(GO:1990879) |
| 0.4 | 1.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.4 | 83.2 | GO:0005764 | lysosome(GO:0005764) |
| 0.3 | 6.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.3 | 5.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.3 | 13.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.3 | 31.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 3.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.2 | 6.1 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.2 | 8.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.2 | 4.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 3.4 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.2 | 6.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 11.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 1.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 22.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 3.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 13.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 13.5 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 32.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 157.7 | GO:0005576 | extracellular region(GO:0005576) |
| 0.1 | 4.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 4.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 14.1 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 28.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 2.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 5.2 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 9.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 12.3 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.8 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 7.0 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 4.9 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 130.8 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 5.2 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 9.8 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.9 | 20.8 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 6.7 | 33.5 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 3.7 | 11.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 2.7 | 23.9 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 2.5 | 17.4 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 2.3 | 13.7 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 1.8 | 12.8 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 1.8 | 12.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 1.8 | 7.0 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 1.7 | 24.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 1.6 | 19.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 1.5 | 9.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 1.5 | 7.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 1.2 | 4.8 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 1.2 | 14.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 1.1 | 24.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 1.1 | 4.3 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 1.0 | 10.6 | GO:0043142 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.9 | 201.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.9 | 6.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.8 | 98.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.8 | 13.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.8 | 4.8 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.8 | 3.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.8 | 6.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.8 | 5.3 | GO:0031779 | melanocortin receptor binding(GO:0031779) |
| 0.7 | 17.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.7 | 5.9 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.7 | 26.4 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.7 | 46.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.7 | 28.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.7 | 6.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.6 | 2.6 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.6 | 7.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.6 | 7.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.6 | 10.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.6 | 8.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.6 | 1.8 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.5 | 25.7 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.5 | 66.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.4 | 2.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.4 | 20.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.4 | 6.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.4 | 4.9 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.4 | 7.9 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 10.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 2.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.3 | 1.7 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.3 | 6.2 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.3 | 2.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.3 | 1.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 4.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.3 | 64.4 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.3 | 1.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.3 | 2.4 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.3 | 3.4 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.3 | 15.6 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.2 | 5.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.2 | 3.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 163.8 | GO:0008289 | lipid binding(GO:0008289) |
| 0.2 | 4.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.2 | 12.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 8.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 10.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 0.8 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 2.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 6.0 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 1.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 14.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 8.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 77.7 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 22.1 | GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen(GO:0016705) |
| 0.1 | 4.8 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 19.6 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 1.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 3.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 18.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 13.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 25.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 6.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 4.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 3.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 6.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 25.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 5.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 5.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 5.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 11.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 1.1 | 14.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.9 | 36.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.7 | 22.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.6 | 19.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.6 | 25.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.6 | 6.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.5 | 73.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.4 | 9.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.4 | 10.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.4 | 30.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.4 | 5.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.4 | 6.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.3 | 8.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 4.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.3 | 7.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.3 | 3.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 6.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.3 | 3.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 28.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.2 | 12.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.2 | 1.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 7.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.2 | 6.0 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.2 | 4.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 6.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 3.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 3.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.4 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 24.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.8 | 22.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 1.6 | 46.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 1.3 | 37.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 1.1 | 11.7 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 1.0 | 13.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 1.0 | 28.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 1.0 | 10.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.9 | 20.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.9 | 5.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.9 | 11.1 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.8 | 10.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.8 | 8.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.8 | 16.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.8 | 10.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.6 | 19.5 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.5 | 4.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.5 | 4.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.5 | 20.7 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.5 | 6.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.5 | 2.4 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.5 | 18.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.5 | 19.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.4 | 7.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.4 | 4.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.4 | 1.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 12.8 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |
| 0.3 | 8.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 12.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 4.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 3.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 7.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 4.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 10.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 4.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 1.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 4.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 6.9 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 2.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.0 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |