Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
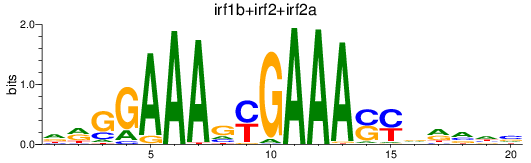
Results for irf1b+irf2+irf2a
Z-value: 3.75
Transcription factors associated with irf1b+irf2+irf2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irf2a
|
ENSDARG00000007387 | interferon regulatory factor 2a |
|
irf2
|
ENSDARG00000040465 | interferon regulatory factor 2 |
|
irf1b
|
ENSDARG00000043249 | interferon regulatory factor 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irf2 | dr11_v1_chr14_-_4121052_4121052 | 0.86 | 1.2e-28 | Click! |
| irf1b | dr11_v1_chr21_+_45627775_45627775 | 0.77 | 1.1e-19 | Click! |
| irf2a | dr11_v1_chr1_+_39865748_39865753 | -0.10 | 3.5e-01 | Click! |
Activity profile of irf1b+irf2+irf2a motif
Sorted Z-values of irf1b+irf2+irf2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_22675475 | 59.33 |
ENSDART00000134423
|
ponzr3
|
plac8 onzin related protein 3 |
| chr19_-_7115229 | 54.45 |
ENSDART00000001930
|
psmb13a
|
proteasome subunit beta 13a |
| chr13_+_13681681 | 48.09 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr19_+_7115223 | 44.64 |
ENSDART00000001359
|
psmb12
|
proteasome subunit beta 12 |
| chr13_+_22675802 | 43.82 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr9_+_30387930 | 40.34 |
ENSDART00000112827
|
si:dkey-18p12.4
|
si:dkey-18p12.4 |
| chr10_-_22095505 | 39.66 |
ENSDART00000140210
|
ponzr10
|
plac8 onzin related protein 10 |
| chr9_-_45601103 | 38.94 |
ENSDART00000180465
|
agr1
|
anterior gradient 1 |
| chr10_+_17776981 | 37.07 |
ENSDART00000141693
|
ccl19b
|
chemokine (C-C motif) ligand 19b |
| chr16_+_23487051 | 35.88 |
ENSDART00000145496
|
icn2
|
ictacalcin 2 |
| chr16_+_29492937 | 34.98 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr3_+_34670076 | 34.12 |
ENSDART00000133457
|
dlx4a
|
distal-less homeobox 4a |
| chr22_-_5553007 | 34.02 |
ENSDART00000165269
|
zgc:171601
|
zgc:171601 |
| chr12_-_4388704 | 33.30 |
ENSDART00000152168
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr1_-_7930679 | 33.14 |
ENSDART00000146090
|
si:dkey-79f11.10
|
si:dkey-79f11.10 |
| chr19_+_1414604 | 31.40 |
ENSDART00000159024
|
btr29
|
bloodthirsty-related gene family, member 29 |
| chr6_-_42983843 | 31.07 |
ENSDART00000130666
|
tnfrsf18
|
tumor necrosis factor receptor superfamily, member 18 |
| chr5_+_13373593 | 30.47 |
ENSDART00000051668
ENSDART00000183883 |
ccl19a.2
|
chemokine (C-C motif) ligand 19a, tandem duplicate 2 |
| chr7_+_22637515 | 30.02 |
ENSDART00000158698
|
si:dkey-112a7.5
|
si:dkey-112a7.5 |
| chr11_+_1584747 | 28.86 |
ENSDART00000154583
|
si:dkey-40c23.2
|
si:dkey-40c23.2 |
| chr13_-_34858500 | 28.44 |
ENSDART00000184843
|
sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr1_+_1805294 | 28.28 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 3 |
| chr23_+_26142807 | 27.29 |
ENSDART00000158878
|
ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr19_+_7124337 | 26.68 |
ENSDART00000031380
|
tap2a
|
transporter associated with antigen processing, subunit type a |
| chr6_-_11091449 | 26.60 |
ENSDART00000127209
|
pttg1ipa
|
PTTG1 interacting protein a |
| chr4_-_17669881 | 26.39 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr5_-_38170996 | 25.55 |
ENSDART00000145805
|
si:ch211-284e13.12
|
si:ch211-284e13.12 |
| chr23_+_26142613 | 25.46 |
ENSDART00000165046
|
ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr11_-_20096018 | 25.46 |
ENSDART00000030420
|
ogfrl2
|
opioid growth factor receptor-like 2 |
| chr24_+_12945803 | 25.04 |
ENSDART00000005105
|
psme1
|
proteasome activator subunit 1 |
| chr5_-_38064065 | 24.53 |
ENSDART00000137181
|
si:dkey-111e8.5
|
si:dkey-111e8.5 |
| chr3_+_36424055 | 24.51 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr5_-_38063585 | 24.41 |
ENSDART00000164947
|
si:dkey-111e8.5
|
si:dkey-111e8.5 |
| chr15_-_47929455 | 24.37 |
ENSDART00000064462
|
psma6l
|
proteasome subunit alpha 6, like |
| chr16_-_51271962 | 23.68 |
ENSDART00000164021
ENSDART00000046420 |
serpinb1l1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 1 |
| chr23_+_642001 | 23.59 |
ENSDART00000030643
ENSDART00000124850 |
irf10
|
interferon regulatory factor 10 |
| chr1_-_25177086 | 23.44 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr13_+_7387822 | 23.19 |
ENSDART00000148240
|
exoc3l4
|
exocyst complex component 3-like 4 |
| chr4_-_26095755 | 23.17 |
ENSDART00000100611
ENSDART00000191266 |
si:ch211-244b2.3
|
si:ch211-244b2.3 |
| chr8_+_47099033 | 23.15 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr1_-_52431220 | 23.02 |
ENSDART00000111256
|
zgc:194101
|
zgc:194101 |
| chr11_+_1608348 | 22.90 |
ENSDART00000162438
|
si:dkey-40c23.3
|
si:dkey-40c23.3 |
| chr4_-_12795030 | 22.50 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr8_-_36469117 | 22.12 |
ENSDART00000111240
|
mhc2dab
|
major histocompatibility complex class II DAB gene |
| chr17_-_23631400 | 22.09 |
ENSDART00000079563
|
fas
|
Fas cell surface death receptor |
| chr11_+_1575435 | 21.65 |
ENSDART00000155713
ENSDART00000156562 |
si:dkey-40c23.1
|
si:dkey-40c23.1 |
| chr21_-_22673758 | 21.62 |
ENSDART00000164910
|
gig2i
|
grass carp reovirus (GCRV)-induced gene 2i |
| chr18_-_38270077 | 21.58 |
ENSDART00000185546
|
caprin1b
|
cell cycle associated protein 1b |
| chr14_+_36889893 | 21.55 |
ENSDART00000124159
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr3_+_29941777 | 21.45 |
ENSDART00000113889
|
ifi35
|
interferon-induced protein 35 |
| chr4_-_26108053 | 21.21 |
ENSDART00000066951
|
si:ch211-244b2.4
|
si:ch211-244b2.4 |
| chr16_+_48753664 | 20.87 |
ENSDART00000155148
|
si:ch73-31d8.2
|
si:ch73-31d8.2 |
| chr18_-_38270596 | 20.77 |
ENSDART00000098889
|
caprin1b
|
cell cycle associated protein 1b |
| chr23_+_642395 | 20.71 |
ENSDART00000186995
|
irf10
|
interferon regulatory factor 10 |
| chr5_-_57723929 | 20.66 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr22_+_29990448 | 20.41 |
ENSDART00000165313
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr13_-_37615029 | 20.22 |
ENSDART00000111199
|
si:dkey-188i13.6
|
si:dkey-188i13.6 |
| chr3_-_22366562 | 20.11 |
ENSDART00000129447
|
ifnphi1
|
interferon phi 1 |
| chr25_+_4751879 | 20.05 |
ENSDART00000169465
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr1_-_58002973 | 19.97 |
ENSDART00000140390
|
si:ch211-114l13.9
|
si:ch211-114l13.9 |
| chr23_-_5101847 | 19.89 |
ENSDART00000122240
|
etv7
|
ets variant 7 |
| chr19_-_15281996 | 19.88 |
ENSDART00000103784
|
edn2
|
endothelin 2 |
| chr22_-_36926342 | 19.87 |
ENSDART00000151804
|
si:dkey-37m8.11
|
si:dkey-37m8.11 |
| chr23_-_37575030 | 19.69 |
ENSDART00000031875
|
tor1l3
|
torsin family 1 like 3 |
| chr7_+_39416336 | 19.32 |
ENSDART00000171783
|
CT030188.1
|
|
| chr5_-_38197080 | 19.14 |
ENSDART00000140708
|
si:ch211-284e13.9
|
si:ch211-284e13.9 |
| chr19_+_23296616 | 18.99 |
ENSDART00000134567
|
irgf1
|
immunity-related GTPase family, f1 |
| chr1_-_8020589 | 18.86 |
ENSDART00000143881
|
si:dkeyp-9d4.2
|
si:dkeyp-9d4.2 |
| chr4_-_26107841 | 18.86 |
ENSDART00000172012
|
si:ch211-244b2.4
|
si:ch211-244b2.4 |
| chr4_+_5842433 | 18.86 |
ENSDART00000124085
ENSDART00000179848 |
usp18
|
ubiquitin specific peptidase 18 |
| chr19_+_7575141 | 18.71 |
ENSDART00000051528
|
s100u
|
S100 calcium binding protein U |
| chr19_+_7575341 | 18.61 |
ENSDART00000134271
|
s100u
|
S100 calcium binding protein U |
| chr2_-_42035250 | 18.39 |
ENSDART00000056460
ENSDART00000140788 |
gbp1
|
guanylate binding protein 1 |
| chr12_+_17106117 | 18.28 |
ENSDART00000149990
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr18_+_17624295 | 18.21 |
ENSDART00000151865
ENSDART00000190408 ENSDART00000189204 ENSDART00000188894 ENSDART00000033762 ENSDART00000193471 |
nlrc5
|
NLR family, CARD domain containing 5 |
| chr20_+_26916639 | 18.08 |
ENSDART00000077787
|
serpinb1l2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 2 |
| chr13_+_47810211 | 17.94 |
ENSDART00000122964
|
zmp:0000001006
|
zmp:0000001006 |
| chr5_-_37116265 | 17.93 |
ENSDART00000057613
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr3_-_22366032 | 17.86 |
ENSDART00000029849
|
ifnphi1
|
interferon phi 1 |
| chr24_-_34680956 | 17.85 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr1_+_58332000 | 17.82 |
ENSDART00000145234
|
ggt1l2.1
|
gamma-glutamyltransferase 1 like 2.1 |
| chr7_+_31891110 | 17.71 |
ENSDART00000173883
|
mybpc3
|
myosin binding protein C, cardiac |
| chr7_+_2259413 | 17.51 |
ENSDART00000173420
|
si:dkey-187j14.7
|
si:dkey-187j14.7 |
| chr22_+_36902352 | 17.37 |
ENSDART00000036273
|
trim35-20
|
tripartite motif containing 35-20 |
| chr16_-_22192006 | 17.25 |
ENSDART00000163338
|
il6r
|
interleukin 6 receptor |
| chr4_-_76383223 | 17.20 |
ENSDART00000161866
ENSDART00000168456 ENSDART00000174287 ENSDART00000174048 |
zgc:123107
|
zgc:123107 |
| chr23_+_46183410 | 17.17 |
ENSDART00000167596
ENSDART00000151149 ENSDART00000150896 |
btr31
|
bloodthirsty-related gene family, member 31 |
| chr22_-_10580194 | 17.16 |
ENSDART00000105848
|
SHISA4 (1 of many)
|
si:dkey-42i9.7 |
| chr3_-_29941357 | 17.10 |
ENSDART00000147732
ENSDART00000137973 ENSDART00000103523 |
grna
|
granulin a |
| chr17_+_25397070 | 17.05 |
ENSDART00000164254
|
zgc:154055
|
zgc:154055 |
| chr3_-_336299 | 17.02 |
ENSDART00000105021
|
mhc1zfa
|
major histocompatibility complex class I ZFA |
| chr5_+_38612134 | 17.01 |
ENSDART00000135600
ENSDART00000181144 |
si:ch211-271e10.2
|
si:ch211-271e10.2 |
| chr13_-_46200240 | 16.90 |
ENSDART00000056984
|
ftr69
|
finTRIM family, member 69 |
| chr23_-_5783421 | 16.87 |
ENSDART00000131521
ENSDART00000019455 |
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr16_-_50952266 | 16.72 |
ENSDART00000165408
|
si:dkeyp-97a10.3
|
si:dkeyp-97a10.3 |
| chr1_-_7951002 | 16.57 |
ENSDART00000138187
|
si:dkey-79f11.8
|
si:dkey-79f11.8 |
| chr15_-_36347858 | 16.34 |
ENSDART00000155274
ENSDART00000157936 |
si:dkey-23k10.2
|
si:dkey-23k10.2 |
| chr23_-_44723102 | 16.32 |
ENSDART00000129138
|
mogat3a
|
monoacylglycerol O-acyltransferase 3a |
| chr3_+_1724941 | 16.17 |
ENSDART00000193402
|
BX321875.2
|
|
| chr15_-_18209672 | 16.16 |
ENSDART00000141508
ENSDART00000136280 |
btr16
|
bloodthirsty-related gene family, member 16 |
| chr12_-_30583668 | 15.95 |
ENSDART00000153406
|
casp7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr6_+_13045885 | 15.90 |
ENSDART00000104757
|
casp8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr18_-_38270430 | 15.84 |
ENSDART00000139519
|
caprin1b
|
cell cycle associated protein 1b |
| chr18_+_30998472 | 15.75 |
ENSDART00000154993
ENSDART00000099333 |
cd151l
|
CD151 antigen, like |
| chr10_+_5060191 | 15.75 |
ENSDART00000145908
ENSDART00000122397 |
carm1l
|
coactivator-associated arginine methyltransferase 1, like |
| chr3_+_24603923 | 15.55 |
ENSDART00000172589
|
si:dkey-68o6.6
|
si:dkey-68o6.6 |
| chr19_-_7144548 | 15.52 |
ENSDART00000147177
ENSDART00000134850 |
psmb8a
psmb13a
|
proteasome subunit beta 8A proteasome subunit beta 13a |
| chr8_-_13641022 | 15.47 |
ENSDART00000080875
|
b3gnt3
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
| chr7_+_20344222 | 15.37 |
ENSDART00000141186
ENSDART00000139274 |
ponzr1
|
plac8 onzin related protein 1 |
| chr11_-_30341431 | 15.33 |
ENSDART00000078378
|
cflara
|
CASP8 and FADD-like apoptosis regulator a |
| chr21_-_7265219 | 15.30 |
ENSDART00000158852
|
egfl7
|
EGF-like-domain, multiple 7 |
| chr7_+_3442834 | 15.15 |
ENSDART00000138247
|
si:ch211-285c6.2
|
si:ch211-285c6.2 |
| chr10_+_7585928 | 15.01 |
ENSDART00000166686
|
tex15
|
testis expressed 15 |
| chr9_+_25330905 | 14.93 |
ENSDART00000101470
|
itm2bb
|
integral membrane protein 2Bb |
| chr24_-_39858710 | 14.72 |
ENSDART00000134251
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr9_+_3153717 | 14.65 |
ENSDART00000191958
|
si:zfos-979f1.2
|
si:zfos-979f1.2 |
| chr4_-_76206413 | 14.64 |
ENSDART00000170738
|
si:ch211-106j21.5
|
si:ch211-106j21.5 |
| chr18_+_13248956 | 14.63 |
ENSDART00000080709
|
plcg2
|
phospholipase C, gamma 2 |
| chr9_+_42269059 | 14.62 |
ENSDART00000113435
|
si:dkey-10c21.1
|
si:dkey-10c21.1 |
| chr16_-_9802449 | 14.61 |
ENSDART00000081208
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr7_+_20505311 | 14.50 |
ENSDART00000187335
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr22_-_18164671 | 14.35 |
ENSDART00000014057
|
rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr21_-_33478164 | 14.32 |
ENSDART00000191542
|
si:ch73-42p12.2
|
si:ch73-42p12.2 |
| chr18_-_14860435 | 14.28 |
ENSDART00000018502
|
mapk12a
|
mitogen-activated protein kinase 12a |
| chr7_+_20344486 | 14.26 |
ENSDART00000134004
ENSDART00000139685 |
ponzr1
|
plac8 onzin related protein 1 |
| chr25_-_31629095 | 14.25 |
ENSDART00000170673
ENSDART00000166930 |
lamb1a
|
laminin, beta 1a |
| chr9_+_3153466 | 14.21 |
ENSDART00000135619
|
si:zfos-979f1.2
|
si:zfos-979f1.2 |
| chr7_-_19614916 | 14.20 |
ENSDART00000169029
|
zgc:194655
|
zgc:194655 |
| chr22_+_5687615 | 14.19 |
ENSDART00000133241
ENSDART00000019854 ENSDART00000138102 |
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr22_-_6941098 | 14.11 |
ENSDART00000105864
|
zgc:171500
|
zgc:171500 |
| chr11_+_27347076 | 14.11 |
ENSDART00000173383
|
fbln2
|
fibulin 2 |
| chr23_-_42752550 | 14.06 |
ENSDART00000187059
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr3_+_1942219 | 14.02 |
ENSDART00000114520
|
zgc:165583
|
zgc:165583 |
| chr7_-_8981507 | 13.91 |
ENSDART00000161422
|
si:ch211-183d5.2
|
si:ch211-183d5.2 |
| chr5_+_22406672 | 13.85 |
ENSDART00000141385
|
si:dkey-27p18.3
|
si:dkey-27p18.3 |
| chr3_+_24618012 | 13.85 |
ENSDART00000111997
|
zgc:171506
|
zgc:171506 |
| chr20_-_34663209 | 13.71 |
ENSDART00000132545
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr3_-_53508580 | 13.70 |
ENSDART00000073978
|
zgc:171711
|
zgc:171711 |
| chr8_-_23240156 | 13.69 |
ENSDART00000131632
|
ptk6a
|
PTK6 protein tyrosine kinase 6a |
| chr3_-_32958505 | 13.52 |
ENSDART00000147374
ENSDART00000136919 |
casp6l1
|
caspase 6, apoptosis-related cysteine peptidase, like 1 |
| chr15_-_41756454 | 13.47 |
ENSDART00000139498
ENSDART00000099302 |
ftr72
|
finTRIM family, member 72 |
| chr2_-_37862380 | 13.46 |
ENSDART00000186005
|
si:ch211-284o19.8
|
si:ch211-284o19.8 |
| chr5_-_30382925 | 13.46 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr3_-_32611833 | 13.39 |
ENSDART00000151282
|
si:ch73-248e21.5
|
si:ch73-248e21.5 |
| chr4_-_77216726 | 13.18 |
ENSDART00000099943
|
psmb10
|
proteasome subunit beta 10 |
| chr2_+_45479841 | 13.18 |
ENSDART00000151856
|
si:ch211-66k16.28
|
si:ch211-66k16.28 |
| chr20_-_1439256 | 13.18 |
ENSDART00000002928
|
themis
|
thymocyte selection associated |
| chr22_-_18164835 | 13.10 |
ENSDART00000143189
|
rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr5_+_38619813 | 13.09 |
ENSDART00000133314
|
si:ch211-271e10.3
|
si:ch211-271e10.3 |
| chr4_-_76079075 | 13.02 |
ENSDART00000158469
|
si:ch211-232d10.3
|
si:ch211-232d10.3 |
| chr11_+_37638873 | 12.97 |
ENSDART00000186384
ENSDART00000184291 ENSDART00000131782 ENSDART00000140502 |
sh2d5
|
SH2 domain containing 5 |
| chr10_+_35358675 | 12.89 |
ENSDART00000193263
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr23_+_43177290 | 12.87 |
ENSDART00000193300
ENSDART00000186065 |
si:dkey-65j6.2
|
si:dkey-65j6.2 |
| chr7_+_19482084 | 12.78 |
ENSDART00000173873
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr3_-_16142057 | 12.77 |
ENSDART00000016616
|
arl5c
|
ADP-ribosylation factor-like 5C |
| chr5_+_4338874 | 12.64 |
ENSDART00000141866
|
sat1a.1
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 1 |
| chr22_+_19290199 | 12.60 |
ENSDART00000148173
|
si:dkey-21e2.15
|
si:dkey-21e2.15 |
| chr22_+_31023205 | 12.58 |
ENSDART00000111561
|
zmp:0000000735
|
zmp:0000000735 |
| chr5_+_40835601 | 12.57 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr12_+_20641471 | 12.56 |
ENSDART00000133654
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr22_+_19289970 | 12.51 |
ENSDART00000137976
ENSDART00000132386 |
si:dkey-21e2.15
|
si:dkey-21e2.15 |
| chr21_-_43428040 | 12.50 |
ENSDART00000148325
|
stk26
|
serine/threonine protein kinase 26 |
| chr25_+_245018 | 12.48 |
ENSDART00000155344
|
zgc:92481
|
zgc:92481 |
| chr5_+_38684651 | 12.45 |
ENSDART00000137852
|
si:dkey-58f10.10
|
si:dkey-58f10.10 |
| chr23_+_5524247 | 12.35 |
ENSDART00000189679
ENSDART00000083622 |
tead3a
|
TEA domain family member 3 a |
| chr15_+_32268790 | 12.33 |
ENSDART00000154457
|
fhdc4
|
FH2 domain containing 4 |
| chr24_+_38216340 | 12.26 |
ENSDART00000188561
|
CU896602.4
|
|
| chr5_-_38820046 | 12.23 |
ENSDART00000182886
|
cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr10_+_8885769 | 12.19 |
ENSDART00000139466
|
itga2.2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 2 |
| chr8_-_30204650 | 12.16 |
ENSDART00000133209
|
zgc:162939
|
zgc:162939 |
| chr22_-_17459587 | 12.12 |
ENSDART00000142267
|
si:ch211-197g15.10
|
si:ch211-197g15.10 |
| chr12_-_37299646 | 12.05 |
ENSDART00000146142
ENSDART00000085201 |
pmp22b
|
peripheral myelin protein 22b |
| chr2_-_8611675 | 12.03 |
ENSDART00000138223
|
si:ch211-71m22.1
|
si:ch211-71m22.1 |
| chr1_-_57839070 | 12.02 |
ENSDART00000152571
|
si:dkey-1c7.3
|
si:dkey-1c7.3 |
| chr18_+_26422124 | 11.99 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr22_+_6254194 | 11.94 |
ENSDART00000112388
ENSDART00000135176 |
rnasel4
|
ribonuclease like 4 |
| chr21_+_8533533 | 11.88 |
ENSDART00000077924
|
FO834888.1
|
|
| chr1_-_8141135 | 11.87 |
ENSDART00000152295
|
FAM83G
|
si:dkeyp-9d4.4 |
| chr3_-_24093144 | 11.85 |
ENSDART00000103982
|
nfe2l1a
|
nuclear factor, erythroid 2-like 1a |
| chr6_+_52947699 | 11.84 |
ENSDART00000180913
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr12_+_17154655 | 11.83 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr22_-_5518117 | 11.81 |
ENSDART00000164613
|
CABZ01064972.1
|
|
| chr4_+_7822773 | 11.63 |
ENSDART00000171391
|
si:ch1073-67j19.2
|
si:ch1073-67j19.2 |
| chr15_-_41734639 | 11.62 |
ENSDART00000154230
ENSDART00000167443 |
ftr90
|
finTRIM family, member 90 |
| chr13_-_42306348 | 11.55 |
ENSDART00000003706
|
kmo
|
kynurenine 3-monooxygenase |
| chr14_+_12169979 | 11.54 |
ENSDART00000129953
|
rhogd
|
ras homolog gene family, member Gd |
| chr8_-_27687095 | 11.50 |
ENSDART00000086946
|
mov10b.1
|
Moloney leukemia virus 10b, tandem duplicate 1 |
| chr3_-_48980319 | 11.44 |
ENSDART00000165319
|
ftr42
|
finTRIM family, member 42 |
| chr9_+_18023288 | 11.42 |
ENSDART00000098355
|
tnfsf11
|
TNF superfamily member 11 |
| chr8_+_25629722 | 11.42 |
ENSDART00000026807
|
wasa
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) a |
| chr15_-_20180475 | 11.40 |
ENSDART00000048183
|
exoc3l2b
|
exocyst complex component 3-like 2b |
| chr7_+_19483277 | 11.39 |
ENSDART00000173750
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr7_+_19615056 | 11.39 |
ENSDART00000124752
ENSDART00000190297 |
si:ch211-212k18.15
|
si:ch211-212k18.15 |
| chr15_-_29162193 | 11.38 |
ENSDART00000138449
ENSDART00000099885 |
xaf1
|
XIAP associated factor 1 |
| chr15_-_18197008 | 11.37 |
ENSDART00000147632
|
si:ch211-247l8.8
|
si:ch211-247l8.8 |
| chr23_-_42752387 | 11.36 |
ENSDART00000149781
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr4_+_60482963 | 11.30 |
ENSDART00000157679
|
si:dkey-211i20.2
|
si:dkey-211i20.2 |
| chr19_+_4066449 | 11.23 |
ENSDART00000162461
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr6_+_36839509 | 11.12 |
ENSDART00000190605
ENSDART00000104160 |
zgc:110788
|
zgc:110788 |
| chr23_+_16864130 | 11.11 |
ENSDART00000060181
|
zgc:114174
|
zgc:114174 |
Network of associatons between targets according to the STRING database.
First level regulatory network of irf1b+irf2+irf2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.0 | 48.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 6.1 | 18.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 5.5 | 22.1 | GO:0070227 | lymphocyte apoptotic process(GO:0070227) |
| 5.3 | 15.9 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) |
| 5.2 | 52.2 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 5.0 | 19.9 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 4.7 | 38.0 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 4.5 | 142.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 4.4 | 13.2 | GO:0043383 | negative T cell selection(GO:0043383) |
| 4.3 | 43.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 3.9 | 19.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 3.7 | 14.9 | GO:0042985 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 3.5 | 17.7 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 3.3 | 26.2 | GO:0031179 | peptide modification(GO:0031179) |
| 3.2 | 12.8 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 3.2 | 12.6 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 3.1 | 40.7 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 3.0 | 9.0 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 2.9 | 17.2 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 2.7 | 13.6 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 2.7 | 29.6 | GO:0072088 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 2.7 | 8.0 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 2.6 | 7.9 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 2.6 | 28.4 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 2.4 | 14.3 | GO:0055016 | hypochord development(GO:0055016) |
| 2.3 | 16.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 2.3 | 11.5 | GO:0043420 | anthranilate metabolic process(GO:0043420) |
| 2.3 | 11.5 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 2.1 | 17.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 2.0 | 9.8 | GO:0035739 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 1.9 | 7.7 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 1.8 | 7.3 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 1.7 | 7.0 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 1.7 | 8.7 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 1.6 | 42.5 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 1.5 | 6.0 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 1.5 | 10.5 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 1.5 | 28.3 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 1.4 | 7.0 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 1.4 | 15.3 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 1.3 | 54.4 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 1.3 | 5.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 1.3 | 3.9 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 1.3 | 8.8 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 1.2 | 22.7 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 1.2 | 15.0 | GO:0007140 | male meiosis(GO:0007140) |
| 1.1 | 5.6 | GO:0019374 | galactosylceramide metabolic process(GO:0006681) galactolipid metabolic process(GO:0019374) |
| 1.1 | 4.3 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 1.1 | 4.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 1.0 | 3.0 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 1.0 | 16.9 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.9 | 14.2 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.9 | 10.2 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.8 | 12.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.8 | 4.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.8 | 18.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.8 | 11.4 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.8 | 9.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.7 | 2.9 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.7 | 4.3 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.7 | 31.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.7 | 9.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.7 | 17.0 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.6 | 14.7 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.6 | 2.9 | GO:0014005 | microglia development(GO:0014005) |
| 0.6 | 16.7 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.6 | 41.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.6 | 58.7 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.5 | 12.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.5 | 4.2 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.5 | 46.4 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.5 | 25.6 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.5 | 8.7 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.5 | 2.0 | GO:0061045 | negative regulation of blood coagulation(GO:0030195) negative regulation of wound healing(GO:0061045) negative regulation of hemostasis(GO:1900047) |
| 0.5 | 25.4 | GO:0010952 | positive regulation of endopeptidase activity(GO:0010950) positive regulation of peptidase activity(GO:0010952) |
| 0.5 | 11.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.5 | 2.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.5 | 27.2 | GO:0045785 | positive regulation of cell adhesion(GO:0045785) |
| 0.4 | 12.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.4 | 11.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.4 | 2.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.4 | 1.3 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.4 | 8.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.4 | 2.4 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.4 | 14.6 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.4 | 8.5 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.4 | 45.8 | GO:0017038 | protein import(GO:0017038) |
| 0.4 | 3.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.4 | 36.9 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.4 | 7.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.4 | 10.2 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.4 | 1.4 | GO:0060220 | blood vessel maturation(GO:0001955) retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.3 | 3.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.3 | 17.0 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.3 | 2.0 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.3 | 18.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.3 | 11.1 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.3 | 3.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.3 | 10.9 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.3 | 1.7 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.3 | 4.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.3 | 8.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.3 | 1.7 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.3 | 34.7 | GO:0021782 | glial cell development(GO:0021782) |
| 0.3 | 3.3 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.3 | 6.0 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.3 | 4.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 3.5 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.2 | 30.7 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.2 | 0.7 | GO:0044036 | cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.2 | 16.9 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.2 | 12.7 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.2 | 11.4 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.2 | 1.4 | GO:0032728 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 28.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.2 | 1.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.2 | 0.9 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 15.3 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.2 | 2.8 | GO:0043248 | proteasome assembly(GO:0043248) chaperone-mediated protein complex assembly(GO:0051131) |
| 0.2 | 0.8 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.2 | 2.5 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 1.3 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) medium-chain fatty acid catabolic process(GO:0051793) |
| 0.2 | 5.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.2 | 3.6 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.2 | 6.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 4.0 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 94.7 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.1 | 0.8 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centriole replication(GO:0046600) negative regulation of centrosome cycle(GO:0046606) negative regulation of organelle assembly(GO:1902116) |
| 0.1 | 2.7 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.1 | 9.3 | GO:0001894 | tissue homeostasis(GO:0001894) |
| 0.1 | 7.1 | GO:0007338 | single fertilization(GO:0007338) fertilization(GO:0009566) |
| 0.1 | 1.1 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 7.9 | GO:0007492 | endoderm development(GO:0007492) |
| 0.1 | 0.2 | GO:0036503 | ERAD pathway(GO:0036503) |
| 0.1 | 3.2 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 10.2 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 8.6 | GO:0006401 | RNA catabolic process(GO:0006401) |
| 0.1 | 27.9 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.1 | 1.9 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) negative regulation of gene silencing(GO:0060969) |
| 0.1 | 1.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 13.7 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.1 | 6.8 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 21.2 | GO:0019941 | modification-dependent protein catabolic process(GO:0019941) modification-dependent macromolecule catabolic process(GO:0043632) |
| 0.1 | 2.8 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 1.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.1 | 3.2 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.1 | 6.8 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.1 | 0.5 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 2.4 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.1 | 0.5 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.1 | 2.3 | GO:0030901 | midbrain development(GO:0030901) |
| 0.1 | 28.2 | GO:1902680 | positive regulation of transcription, DNA-templated(GO:0045893) positive regulation of RNA biosynthetic process(GO:1902680) positive regulation of nucleic acid-templated transcription(GO:1903508) |
| 0.0 | 7.2 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 0.4 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 7.1 | GO:0042127 | regulation of cell proliferation(GO:0042127) |
| 0.0 | 5.6 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 6.3 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 2.6 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 4.0 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 3.7 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 84.4 | GO:0035556 | intracellular signal transduction(GO:0035556) |
| 0.0 | 1.5 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 2.1 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 0.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 1.5 | GO:0048884 | neuromast development(GO:0048884) |
| 0.0 | 0.4 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 1.6 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 16.7 | GO:0002376 | immune system process(GO:0002376) |
| 0.0 | 2.7 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
| 0.0 | 1.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 25.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 5.1 | 142.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 4.9 | 39.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 4.2 | 25.5 | GO:0042612 | MHC protein complex(GO:0042611) MHC class I protein complex(GO:0042612) |
| 2.9 | 8.6 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 2.8 | 28.4 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 1.5 | 7.7 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 1.4 | 38.3 | GO:0000145 | exocyst(GO:0000145) |
| 1.3 | 5.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 1.3 | 6.3 | GO:0097433 | dense body(GO:0097433) |
| 1.2 | 6.0 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 1.1 | 18.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 1.0 | 4.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 1.0 | 4.8 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.9 | 17.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.9 | 4.6 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.9 | 4.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.6 | 14.3 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.6 | 4.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.6 | 4.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.5 | 12.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.5 | 23.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.5 | 24.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.4 | 123.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.4 | 3.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.3 | 11.5 | GO:0043186 | P granule(GO:0043186) |
| 0.3 | 2.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.3 | 16.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.3 | 86.9 | GO:0005764 | lysosome(GO:0005764) |
| 0.3 | 2.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.3 | 1.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.3 | 12.8 | GO:0031984 | trans-Golgi network(GO:0005802) organelle subcompartment(GO:0031984) Golgi subcompartment(GO:0098791) |
| 0.3 | 12.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 27.7 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.2 | 3.2 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.2 | 58.9 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.2 | 1.5 | GO:0016586 | RSC complex(GO:0016586) |
| 0.2 | 13.0 | GO:0060076 | postsynaptic density(GO:0014069) excitatory synapse(GO:0060076) |
| 0.2 | 205.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.2 | 0.5 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.2 | 2.6 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 4.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 2.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 33.1 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 2.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 7.9 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.1 | 5.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 15.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 9.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 7.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 6.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 0.6 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 72.6 | GO:0005829 | cytosol(GO:0005829) |
| 0.1 | 3.0 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 3.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 12.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 9.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0032806 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) carboxy-terminal domain protein kinase complex(GO:0032806) |
| 0.0 | 0.7 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 66.6 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 6.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.7 | 38.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 10.6 | 53.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 10.6 | 52.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 6.8 | 20.3 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) |
| 6.2 | 118.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 5.0 | 19.9 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 4.2 | 25.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 4.1 | 28.4 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 3.8 | 11.5 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 3.2 | 47.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 3.2 | 22.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 3.0 | 9.0 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 2.9 | 26.2 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 2.9 | 8.6 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 2.8 | 28.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 2.4 | 65.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 2.3 | 15.8 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 2.2 | 6.6 | GO:0031956 | medium-chain fatty acid-CoA ligase activity(GO:0031956) butyrate-CoA ligase activity(GO:0047760) |
| 2.0 | 6.0 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 1.8 | 93.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 1.8 | 12.6 | GO:0019809 | spermidine binding(GO:0019809) |
| 1.7 | 8.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 1.7 | 6.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 1.6 | 16.3 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 1.6 | 17.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.6 | 61.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 1.6 | 9.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.5 | 4.6 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 1.5 | 10.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 1.4 | 14.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 1.4 | 5.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.3 | 3.9 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 1.3 | 6.3 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 1.2 | 19.7 | GO:0031386 | protein tag(GO:0031386) |
| 1.2 | 54.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 1.2 | 6.0 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) peptide-transporting ATPase activity(GO:0015440) TAP binding(GO:0046977) |
| 1.2 | 4.7 | GO:0031005 | filamin binding(GO:0031005) |
| 1.2 | 11.5 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 1.1 | 4.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 1.1 | 10.8 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 1.0 | 10.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 1.0 | 4.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 1.0 | 12.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 1.0 | 9.8 | GO:0016936 | galactoside binding(GO:0016936) |
| 1.0 | 8.8 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.9 | 19.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.9 | 2.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.8 | 8.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.8 | 12.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.6 | 17.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.6 | 17.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.6 | 2.5 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.6 | 9.8 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.6 | 17.0 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.6 | 7.3 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.6 | 14.7 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.5 | 2.0 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.5 | 16.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.5 | 69.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.5 | 3.4 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.5 | 7.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.5 | 3.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.5 | 5.9 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.4 | 15.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.4 | 11.5 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.4 | 2.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.4 | 7.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.4 | 3.9 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.4 | 4.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.4 | 1.5 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.4 | 13.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.4 | 5.8 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.3 | 5.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.3 | 17.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.3 | 74.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.3 | 35.2 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.3 | 11.0 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.3 | 2.9 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.3 | 4.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.3 | 1.1 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.3 | 4.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.3 | 3.5 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.3 | 8.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.3 | 1.8 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.3 | 13.7 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.3 | 6.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 29.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 0.8 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 3.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 5.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 26.2 | GO:0043492 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.2 | 4.0 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.2 | 177.6 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.2 | 8.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 97.2 | GO:0005525 | GTP binding(GO:0005525) |
| 0.2 | 12.6 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 1.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 7.7 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 0.7 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 4.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 20.8 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 24.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 1.1 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.1 | 2.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 18.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 3.5 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.1 | 2.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.3 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 5.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 2.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 5.4 | GO:0004175 | endopeptidase activity(GO:0004175) |
| 0.0 | 5.6 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 4.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 2.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 8.5 | GO:0016829 | lyase activity(GO:0016829) |
| 0.0 | 1.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 3.5 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 3.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.7 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 5.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 5.2 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 2.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 7.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 9.0 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 2.6 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 38.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 2.2 | 17.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 2.2 | 17.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 1.7 | 53.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 1.0 | 16.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.9 | 19.9 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.7 | 23.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.7 | 17.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.6 | 11.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.4 | 11.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.4 | 8.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.4 | 7.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.4 | 1.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.3 | 18.3 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.2 | 27.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 6.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 3.5 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.2 | 5.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 5.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 10.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 4.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 8.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 3.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 7.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 2.5 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 12.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 0.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 2.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 38.0 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 4.5 | 22.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 4.4 | 56.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 3.5 | 42.0 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 2.6 | 75.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 2.5 | 17.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 2.4 | 24.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 2.3 | 16.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 1.6 | 9.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 1.5 | 44.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 1.5 | 17.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.8 | 18.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.7 | 9.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.7 | 27.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.6 | 15.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.5 | 4.9 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.5 | 3.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.5 | 14.6 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.5 | 5.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.4 | 10.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.4 | 4.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.4 | 17.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.3 | 15.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.3 | 10.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 4.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.3 | 4.3 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.3 | 1.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.3 | 12.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 2.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 0.4 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.2 | 12.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.2 | 3.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 3.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 7.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 3.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 2.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 2.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 0.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 0.8 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 1.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 2.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |