Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
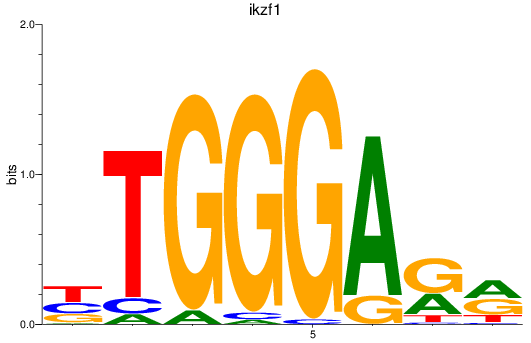
Results for ikzf1
Z-value: 1.32
Transcription factors associated with ikzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ikzf1
|
ENSDARG00000013539 | IKAROS family zinc finger 1 (Ikaros) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ikzf1 | dr11_v1_chr13_-_15986871_15986871 | 0.19 | 6.6e-02 | Click! |
Activity profile of ikzf1 motif
Sorted Z-values of ikzf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_32817274 | 22.05 |
ENSDART00000142582
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr5_-_71722257 | 19.11 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr5_+_32222303 | 17.73 |
ENSDART00000051362
|
myhc4
|
myosin heavy chain 4 |
| chr25_+_29161609 | 17.19 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr6_-_29195642 | 13.91 |
ENSDART00000078625
|
dpt
|
dermatopontin |
| chr23_-_23256726 | 13.65 |
ENSDART00000131353
|
si:dkey-98j1.5
|
si:dkey-98j1.5 |
| chr6_-_39764995 | 12.44 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr7_+_39386982 | 12.36 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr8_-_18535822 | 10.53 |
ENSDART00000100558
|
nexn
|
nexilin (F actin binding protein) |
| chr22_-_29191152 | 10.42 |
ENSDART00000132702
|
pvalb7
|
parvalbumin 7 |
| chr22_-_237651 | 9.89 |
ENSDART00000075210
|
zgc:66156
|
zgc:66156 |
| chr25_-_31396479 | 9.47 |
ENSDART00000156828
|
prr33
|
proline rich 33 |
| chr22_-_282498 | 8.96 |
ENSDART00000182766
|
CABZ01079178.1
|
|
| chr2_-_15324837 | 8.23 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr23_+_6077503 | 7.38 |
ENSDART00000081714
ENSDART00000139834 |
mybpha
|
myosin binding protein Ha |
| chr2_+_25278107 | 7.29 |
ENSDART00000131977
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr13_-_2189761 | 7.23 |
ENSDART00000166255
|
mlip
|
muscular LMNA-interacting protein |
| chr3_-_52614747 | 6.89 |
ENSDART00000154365
|
trim35-13
|
tripartite motif containing 35-13 |
| chr9_-_21067971 | 6.87 |
ENSDART00000004333
|
tbx15
|
T-box 15 |
| chr19_-_38539670 | 6.75 |
ENSDART00000136775
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr2_-_21335131 | 6.67 |
ENSDART00000057022
|
klhl40a
|
kelch-like family member 40a |
| chr24_+_35947077 | 6.27 |
ENSDART00000173406
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr17_+_2549503 | 5.86 |
ENSDART00000156843
|
si:dkey-248g15.3
|
si:dkey-248g15.3 |
| chr15_-_2640966 | 5.66 |
ENSDART00000063320
|
cldne
|
claudin e |
| chr1_+_1838164 | 5.57 |
ENSDART00000006013
|
atp1a1a.5
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 5 |
| chr1_-_59116617 | 5.55 |
ENSDART00000137471
ENSDART00000140490 |
MFAP4 (1 of many)
|
si:zfos-2330d3.7 |
| chr7_+_31879649 | 5.48 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr13_+_24842857 | 5.46 |
ENSDART00000123866
|
dusp13a
|
dual specificity phosphatase 13a |
| chr8_+_1009831 | 5.36 |
ENSDART00000172414
|
fabp1b.2
|
fatty acid binding protein 1b, liver, tandem duplicate 2 |
| chr20_+_15982482 | 5.32 |
ENSDART00000020999
|
angptl1a
|
angiopoietin-like 1a |
| chr23_+_36083529 | 5.27 |
ENSDART00000053295
ENSDART00000130260 |
hoxc10a
|
homeobox C10a |
| chr21_-_131236 | 5.06 |
ENSDART00000160005
|
si:ch1073-398f15.1
|
si:ch1073-398f15.1 |
| chr5_-_57641257 | 5.01 |
ENSDART00000149282
|
hspb2
|
heat shock protein, alpha-crystallin-related, b2 |
| chr20_+_27194833 | 5.00 |
ENSDART00000150072
|
si:dkey-85n7.8
|
si:dkey-85n7.8 |
| chr1_-_10914523 | 4.91 |
ENSDART00000007013
|
dmd
|
dystrophin |
| chr12_-_26430507 | 4.78 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr7_-_35432901 | 4.69 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr18_+_20494413 | 4.67 |
ENSDART00000060295
|
rapsn
|
receptor-associated protein of the synapse, 43kD |
| chr11_+_10541258 | 4.61 |
ENSDART00000132365
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr11_-_21030070 | 4.51 |
ENSDART00000186322
|
fmoda
|
fibromodulin a |
| chr13_-_13754091 | 4.34 |
ENSDART00000131255
|
ky
|
kyphoscoliosis peptidase |
| chr17_+_23462972 | 4.31 |
ENSDART00000112959
ENSDART00000192168 |
ankrd1a
|
ankyrin repeat domain 1a (cardiac muscle) |
| chr7_-_2039060 | 4.22 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr17_-_14671098 | 4.20 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr6_+_18142623 | 4.17 |
ENSDART00000169431
ENSDART00000158841 |
si:dkey-237i9.8
|
si:dkey-237i9.8 |
| chr3_+_30921246 | 4.15 |
ENSDART00000076850
|
cldni
|
claudin i |
| chr6_-_30485009 | 4.11 |
ENSDART00000025698
|
zgc:153311
|
zgc:153311 |
| chr12_-_26415499 | 4.09 |
ENSDART00000185779
|
synpo2lb
|
synaptopodin 2-like b |
| chr18_-_41375120 | 4.05 |
ENSDART00000098673
|
ptx3a
|
pentraxin 3, long a |
| chr9_-_21067673 | 3.98 |
ENSDART00000180257
|
tbx15
|
T-box 15 |
| chr5_+_37517800 | 3.87 |
ENSDART00000048107
|
FP102018.1
|
Danio rerio latent transforming growth factor beta binding protein 3 (ltbp3), mRNA. |
| chr19_-_9648542 | 3.84 |
ENSDART00000172628
|
clcn1a
|
chloride channel, voltage-sensitive 1a |
| chr4_-_4119396 | 3.77 |
ENSDART00000067409
ENSDART00000138221 |
lmod2b
|
leiomodin 2 (cardiac) b |
| chr21_-_35534401 | 3.76 |
ENSDART00000112308
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr5_-_41831646 | 3.71 |
ENSDART00000134326
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr6_-_40899618 | 3.63 |
ENSDART00000153949
ENSDART00000021969 |
zgc:172271
|
zgc:172271 |
| chr2_+_35993404 | 3.60 |
ENSDART00000170845
|
lamc2
|
laminin, gamma 2 |
| chr12_-_47782623 | 3.59 |
ENSDART00000115742
|
selenou1b
|
selenoprotein U1b |
| chr15_+_42573909 | 3.50 |
ENSDART00000181801
|
CLDN8 (1 of many)
|
zgc:110333 |
| chr3_+_49021079 | 3.45 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr17_-_45370200 | 3.40 |
ENSDART00000186208
|
znf106a
|
zinc finger protein 106a |
| chr1_+_51496862 | 3.39 |
ENSDART00000150433
|
meis1a
|
Meis homeobox 1 a |
| chr3_-_59981162 | 3.35 |
ENSDART00000128790
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr21_-_40835069 | 3.30 |
ENSDART00000004686
|
limk1b
|
LIM domain kinase 1b |
| chr5_-_31712399 | 3.28 |
ENSDART00000141328
|
pip5kl1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chr20_+_54738210 | 3.23 |
ENSDART00000151399
|
pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr2_+_55665322 | 3.18 |
ENSDART00000183636
ENSDART00000183814 |
klf2b
|
Kruppel-like factor 2b |
| chr17_-_14876758 | 3.16 |
ENSDART00000155857
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr17_+_45607580 | 3.14 |
ENSDART00000122583
|
si:ch211-202f3.4
|
si:ch211-202f3.4 |
| chr12_+_20641102 | 3.14 |
ENSDART00000152964
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr19_+_22216778 | 3.05 |
ENSDART00000052521
|
nfatc1
|
nuclear factor of activated T cells 1 |
| chr17_+_996509 | 3.00 |
ENSDART00000158830
|
cyp1c2
|
cytochrome P450, family 1, subfamily C, polypeptide 2 |
| chr16_+_20915319 | 3.00 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr5_-_28606916 | 3.00 |
ENSDART00000026107
ENSDART00000137717 |
tnc
|
tenascin C |
| chr24_-_38816725 | 2.97 |
ENSDART00000063231
|
nog2
|
noggin 2 |
| chr2_-_689047 | 2.93 |
ENSDART00000122732
|
foxc1a
|
forkhead box C1a |
| chr15_-_23376541 | 2.92 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr3_-_33967767 | 2.84 |
ENSDART00000151493
ENSDART00000151160 |
ighv1-4
|
immunoglobulin heavy variable 1-4 |
| chr12_+_17042754 | 2.83 |
ENSDART00000066439
|
ch25h
|
cholesterol 25-hydroxylase |
| chr4_-_12914163 | 2.81 |
ENSDART00000140002
ENSDART00000145917 ENSDART00000141355 ENSDART00000067135 |
msrb3
|
methionine sulfoxide reductase B3 |
| chr21_+_13383413 | 2.78 |
ENSDART00000151345
|
zgc:113162
|
zgc:113162 |
| chr15_-_2652640 | 2.75 |
ENSDART00000146094
|
cldnf
|
claudin f |
| chr7_+_39410180 | 2.73 |
ENSDART00000168641
|
CT030188.1
|
|
| chr8_+_44475793 | 2.72 |
ENSDART00000190118
|
si:ch73-211l2.3
|
si:ch73-211l2.3 |
| chr8_-_52229462 | 2.71 |
ENSDART00000185949
ENSDART00000051825 |
tcf7l1b
|
transcription factor 7 like 1b |
| chr3_-_59981476 | 2.69 |
ENSDART00000035878
ENSDART00000124038 |
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr7_+_17816470 | 2.67 |
ENSDART00000173807
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr9_-_34260214 | 2.67 |
ENSDART00000012385
|
me3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr21_+_19525141 | 2.66 |
ENSDART00000058489
|
gzma
|
granzyme A |
| chr5_+_54938634 | 2.61 |
ENSDART00000163050
ENSDART00000145765 |
cd180
|
CD180 molecule |
| chr8_+_52642869 | 2.59 |
ENSDART00000163617
ENSDART00000189997 |
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr22_+_15898221 | 2.58 |
ENSDART00000062587
|
klf2a
|
Kruppel-like factor 2a |
| chr5_-_26181863 | 2.55 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr5_-_41841892 | 2.55 |
ENSDART00000167089
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr1_-_59104145 | 2.53 |
ENSDART00000132495
ENSDART00000152457 |
MFAP4 (1 of many)
si:zfos-2330d3.7
|
si:zfos-2330d3.1 si:zfos-2330d3.7 |
| chr25_-_23526058 | 2.50 |
ENSDART00000191331
ENSDART00000062930 |
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr18_-_21177674 | 2.49 |
ENSDART00000060175
|
si:dkey-12e7.4
|
si:dkey-12e7.4 |
| chr2_+_55665095 | 2.49 |
ENSDART00000059188
|
klf2b
|
Kruppel-like factor 2b |
| chr17_-_33416020 | 2.48 |
ENSDART00000140149
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr7_+_17816006 | 2.47 |
ENSDART00000080834
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr19_+_24872159 | 2.46 |
ENSDART00000158490
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr7_-_58098814 | 2.45 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr15_-_34458495 | 2.44 |
ENSDART00000059954
|
meox2a
|
mesenchyme homeobox 2a |
| chr12_-_8070969 | 2.43 |
ENSDART00000020995
|
tmem26b
|
transmembrane protein 26b |
| chr5_-_41841675 | 2.39 |
ENSDART00000141683
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr5_+_30741730 | 2.39 |
ENSDART00000098246
ENSDART00000186992 ENSDART00000182533 |
ftr83
|
finTRIM family, member 83 |
| chr15_-_33933790 | 2.38 |
ENSDART00000165162
ENSDART00000182258 ENSDART00000183240 |
mag
|
myelin associated glycoprotein |
| chr10_-_8358396 | 2.37 |
ENSDART00000059322
|
csgalnact1a
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1a |
| chr17_+_10242166 | 2.33 |
ENSDART00000170420
|
clec14a
|
C-type lectin domain containing 14A |
| chr3_-_58644920 | 2.33 |
ENSDART00000155953
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr5_-_41838354 | 2.32 |
ENSDART00000146793
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr3_+_32443395 | 2.32 |
ENSDART00000188447
|
prr12b
|
proline rich 12b |
| chr23_+_34047413 | 2.32 |
ENSDART00000143933
ENSDART00000123925 ENSDART00000176139 |
chchd6a
|
coiled-coil-helix-coiled-coil-helix domain containing 6a |
| chr18_-_16792561 | 2.32 |
ENSDART00000145546
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr7_-_24390879 | 2.30 |
ENSDART00000036680
|
ptgr1
|
prostaglandin reductase 1 |
| chr18_+_20869923 | 2.26 |
ENSDART00000138471
|
pgpep1l
|
pyroglutamyl-peptidase I-like |
| chr1_-_206208 | 2.24 |
ENSDART00000060968
|
adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr10_+_31951338 | 2.23 |
ENSDART00000019416
|
lhfpl6
|
LHFPL tetraspan subfamily member 6 |
| chr1_-_26702930 | 2.21 |
ENSDART00000109297
ENSDART00000152389 |
foxe1
|
forkhead box E1 |
| chr7_+_12950507 | 2.21 |
ENSDART00000067629
ENSDART00000158004 |
saa
|
serum amyloid A |
| chr7_+_39410393 | 2.16 |
ENSDART00000158561
ENSDART00000185173 |
CT030188.1
|
|
| chr24_-_25184553 | 2.15 |
ENSDART00000166917
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr21_-_25741096 | 2.14 |
ENSDART00000181756
|
cldnh
|
claudin h |
| chr22_+_30543437 | 2.14 |
ENSDART00000137983
|
si:dkey-103k4.1
|
si:dkey-103k4.1 |
| chr17_+_25414033 | 2.11 |
ENSDART00000001691
|
tdh2
|
L-threonine dehydrogenase 2 |
| chr14_-_5678457 | 2.10 |
ENSDART00000012116
|
tlx2
|
T cell leukemia homeobox 2 |
| chr7_+_27317174 | 2.09 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr3_+_4346854 | 2.05 |
ENSDART00000004273
|
si:dkey-73p2.3
|
si:dkey-73p2.3 |
| chr23_+_21663631 | 2.00 |
ENSDART00000066125
|
dhrs3a
|
dehydrogenase/reductase (SDR family) member 3a |
| chr1_+_26099250 | 1.99 |
ENSDART00000054205
|
ndufb6
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6 |
| chr10_-_8129175 | 1.97 |
ENSDART00000133921
|
ndufa9b
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9b |
| chr19_-_7291733 | 1.96 |
ENSDART00000015559
|
sdha
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr12_+_42574148 | 1.95 |
ENSDART00000157855
|
ebf3a
|
early B cell factor 3a |
| chr19_+_12583577 | 1.94 |
ENSDART00000151508
|
ldlrad4a
|
low density lipoprotein receptor class A domain containing 4a |
| chr6_-_49170390 | 1.91 |
ENSDART00000182255
|
ngfb
|
nerve growth factor b (beta polypeptide) |
| chr10_+_9595575 | 1.90 |
ENSDART00000091780
ENSDART00000184287 |
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr17_+_44697604 | 1.90 |
ENSDART00000156625
|
pgfb
|
placental growth factor b |
| chr14_+_16813816 | 1.89 |
ENSDART00000161201
|
limch1b
|
LIM and calponin homology domains 1b |
| chr15_-_33904831 | 1.89 |
ENSDART00000164333
ENSDART00000165404 |
mag
|
myelin associated glycoprotein |
| chr20_-_27330383 | 1.88 |
ENSDART00000153277
|
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr7_+_2228276 | 1.86 |
ENSDART00000064294
|
si:dkey-187j14.4
|
si:dkey-187j14.4 |
| chr25_+_35304903 | 1.86 |
ENSDART00000034313
ENSDART00000187659 |
gas2a
|
growth arrest-specific 2a |
| chr23_-_20002459 | 1.85 |
ENSDART00000163396
|
b4galt3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr9_+_2507526 | 1.85 |
ENSDART00000166579
|
wipf1a
|
WAS/WASL interacting protein family, member 1a |
| chr12_-_684200 | 1.85 |
ENSDART00000152122
|
si:ch211-176g6.2
|
si:ch211-176g6.2 |
| chr17_+_33415319 | 1.84 |
ENSDART00000140805
ENSDART00000025501 ENSDART00000146447 |
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr14_+_7048930 | 1.84 |
ENSDART00000109138
|
hbegfa
|
heparin-binding EGF-like growth factor a |
| chr3_+_30980852 | 1.82 |
ENSDART00000028529
|
prf1.1
|
perforin 1.1 |
| chr1_+_5275811 | 1.78 |
ENSDART00000189676
|
spry2
|
sprouty RTK signaling antagonist 2 |
| chr7_-_2143313 | 1.78 |
ENSDART00000173871
|
si:cabz01007807.1
|
si:cabz01007807.1 |
| chr21_-_10773344 | 1.77 |
ENSDART00000063244
|
grp
|
gastrin-releasing peptide |
| chr9_+_33145522 | 1.76 |
ENSDART00000005879
|
atp5po
|
ATP synthase peripheral stalk subunit OSCP |
| chr24_-_3304664 | 1.75 |
ENSDART00000183168
|
sugct
|
succinyl-CoA:glutarate-CoA transferase |
| chr21_+_11560153 | 1.74 |
ENSDART00000065842
|
cd8a
|
CD8a molecule |
| chr3_+_41922114 | 1.74 |
ENSDART00000138280
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr7_-_43787616 | 1.73 |
ENSDART00000179758
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr7_-_3807658 | 1.73 |
ENSDART00000126799
ENSDART00000104372 |
zgc:195077
|
zgc:195077 |
| chr12_-_30548244 | 1.72 |
ENSDART00000193616
|
zgc:158404
|
zgc:158404 |
| chr11_+_6456146 | 1.72 |
ENSDART00000036939
|
gadd45ba
|
growth arrest and DNA-damage-inducible, beta a |
| chr20_-_34292607 | 1.70 |
ENSDART00000144705
|
hmcn1
|
hemicentin 1 |
| chr21_+_42713845 | 1.70 |
ENSDART00000166927
|
si:ch1073-204b8.1
|
si:ch1073-204b8.1 |
| chr9_-_54304684 | 1.69 |
ENSDART00000109512
|
il13
|
interleukin 13 |
| chr17_+_33415542 | 1.69 |
ENSDART00000183169
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr13_+_25412307 | 1.68 |
ENSDART00000175339
|
calhm1
|
calcium homeostasis modulator 1 |
| chr20_+_33534038 | 1.65 |
ENSDART00000029206
|
kcnf1a
|
potassium voltage-gated channel, subfamily F, member 1a |
| chr21_+_13387965 | 1.65 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr16_-_42151909 | 1.64 |
ENSDART00000160950
|
dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr22_+_12431608 | 1.63 |
ENSDART00000108609
|
rnd3a
|
Rho family GTPase 3a |
| chr15_-_33925851 | 1.62 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr2_+_3428357 | 1.61 |
ENSDART00000125967
|
CU633991.1
|
|
| chr5_+_20437124 | 1.59 |
ENSDART00000159001
|
tmem119a
|
transmembrane protein 119a |
| chr25_-_20238793 | 1.59 |
ENSDART00000145987
|
dnm1l
|
dynamin 1-like |
| chr19_-_40199081 | 1.59 |
ENSDART00000051970
ENSDART00000151079 |
grn2
|
granulin 2 |
| chr10_+_35347993 | 1.58 |
ENSDART00000131350
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr6_-_19168640 | 1.58 |
ENSDART00000163183
|
si:ch211-188c18.1
|
si:ch211-188c18.1 |
| chr13_-_36391496 | 1.58 |
ENSDART00000100217
ENSDART00000140243 |
actn1
|
actinin, alpha 1 |
| chr3_-_3209432 | 1.57 |
ENSDART00000140635
|
si:ch211-229i14.2
|
si:ch211-229i14.2 |
| chr7_+_34549377 | 1.56 |
ENSDART00000191814
|
fhod1
|
formin homology 2 domain containing 1 |
| chr2_-_50053006 | 1.54 |
ENSDART00000083654
|
si:ch211-106n13.3
|
si:ch211-106n13.3 |
| chr20_-_34292285 | 1.53 |
ENSDART00000020389
|
hmcn1
|
hemicentin 1 |
| chr8_-_51579286 | 1.52 |
ENSDART00000147878
|
ankrd39
|
ankyrin repeat domain 39 |
| chr7_+_17816623 | 1.51 |
ENSDART00000173963
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr22_-_15693085 | 1.46 |
ENSDART00000141861
|
si:ch1073-396h14.1
|
si:ch1073-396h14.1 |
| chr12_+_23762966 | 1.45 |
ENSDART00000152942
ENSDART00000181725 |
jcada
|
junctional cadherin 5 associated a |
| chr14_-_3268155 | 1.44 |
ENSDART00000177244
|
pdgfrb
|
platelet-derived growth factor receptor, beta polypeptide |
| chr3_-_34084387 | 1.44 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr2_-_2642476 | 1.44 |
ENSDART00000124032
|
serbp1b
|
SERPINE1 mRNA binding protein 1b |
| chr7_-_20731078 | 1.43 |
ENSDART00000188267
|
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr24_-_2900511 | 1.42 |
ENSDART00000185674
|
fam69c
|
family with sequence similarity 69, member C |
| chr9_-_1939232 | 1.42 |
ENSDART00000146131
|
hoxd3a
|
homeobox D3a |
| chr14_+_26229056 | 1.41 |
ENSDART00000179045
|
LO018208.1
|
|
| chr6_-_54107269 | 1.41 |
ENSDART00000190017
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr16_-_51299061 | 1.39 |
ENSDART00000148677
|
serpinb1l4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 4 |
| chr2_+_6885852 | 1.38 |
ENSDART00000016607
|
rgs5b
|
regulator of G protein signaling 5b |
| chr25_-_19224298 | 1.37 |
ENSDART00000149917
|
acanb
|
aggrecan b |
| chr17_-_23412705 | 1.37 |
ENSDART00000126995
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr13_+_52061034 | 1.34 |
ENSDART00000170383
|
CABZ01089777.1
|
|
| chr2_+_56012016 | 1.33 |
ENSDART00000146160
ENSDART00000188702 |
loxl5b
|
lysyl oxidase-like 5b |
| chr15_-_47193564 | 1.33 |
ENSDART00000172453
|
LSAMP
|
limbic system-associated membrane protein |
| chr23_+_642001 | 1.32 |
ENSDART00000030643
ENSDART00000124850 |
irf10
|
interferon regulatory factor 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ikzf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.2 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 2.7 | 19.1 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 2.2 | 6.7 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 1.5 | 10.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 1.1 | 12.4 | GO:0061718 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 1.1 | 5.5 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 1.0 | 3.0 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.7 | 2.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.7 | 2.9 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.7 | 2.8 | GO:0030091 | protein repair(GO:0030091) |
| 0.7 | 2.0 | GO:1901006 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.5 | 2.7 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.5 | 1.6 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.5 | 14.5 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.5 | 2.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.4 | 1.8 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.4 | 2.6 | GO:0055016 | hypochord development(GO:0055016) |
| 0.4 | 3.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.4 | 1.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.4 | 4.8 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.4 | 2.0 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.4 | 1.5 | GO:1904355 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.4 | 2.2 | GO:0051883 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.4 | 1.4 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.4 | 1.4 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.3 | 1.0 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.3 | 3.0 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.3 | 11.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.3 | 5.6 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.3 | 4.7 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.3 | 0.6 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.3 | 1.9 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.3 | 1.9 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.3 | 1.3 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.3 | 2.1 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.3 | 13.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.3 | 2.6 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.3 | 0.8 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.3 | 1.0 | GO:0010712 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.2 | 1.7 | GO:1904019 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.2 | 1.0 | GO:0014060 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.2 | 1.6 | GO:0032231 | regulation of actin filament bundle assembly(GO:0032231) |
| 0.2 | 1.1 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.2 | 0.9 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.2 | 0.9 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.2 | 1.3 | GO:0055014 | outflow tract morphogenesis(GO:0003151) atrial cardiac muscle cell development(GO:0055014) |
| 0.2 | 2.1 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.2 | 1.8 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.2 | 3.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.2 | 9.8 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.2 | 3.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 0.6 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.2 | 1.1 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.2 | 3.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.2 | 1.8 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.2 | 1.9 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.2 | 12.5 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.2 | 2.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 1.9 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 2.2 | GO:0098868 | endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.2 | 2.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 1.2 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.2 | 1.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 2.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 2.0 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 4.6 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.2 | 0.5 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.1 | 2.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.9 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 2.3 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.1 | 7.3 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.9 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 0.8 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.5 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.9 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 3.5 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 2.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 5.0 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.1 | 1.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 3.0 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.1 | 0.4 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.1 | 0.7 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.9 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 6.3 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 0.4 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 0.4 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.6 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.1 | 3.2 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.1 | 3.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 4.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 1.0 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 5.5 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 2.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.4 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 1.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 8.2 | GO:0001708 | cell fate specification(GO:0001708) |
| 0.1 | 1.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 4.0 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 1.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 1.6 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.1 | 0.8 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 1.9 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 0.6 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.9 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 1.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 3.9 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.1 | 1.5 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 1.8 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 4.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.3 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 0.1 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.0 | 0.4 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 3.3 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.7 | GO:0098943 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 1.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 1.9 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.4 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.9 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 8.2 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 1.1 | GO:0001935 | endothelial cell proliferation(GO:0001935) |
| 0.0 | 1.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.1 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.2 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.0 | 0.7 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.5 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 1.0 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.0 | 1.8 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 1.1 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.6 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.4 | GO:0035924 | cellular response to vascular endothelial growth factor stimulus(GO:0035924) |
| 0.0 | 0.1 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.5 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 1.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.3 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.7 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.2 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.2 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.9 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.3 | GO:1901653 | cellular response to peptide hormone stimulus(GO:0071375) cellular response to peptide(GO:1901653) |
| 0.0 | 0.8 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.0 | 3.4 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.1 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 2.2 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 2.4 | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules(GO:0098742) |
| 0.0 | 0.2 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 12.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.5 | 1.9 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.4 | 12.9 | GO:0031672 | A band(GO:0031672) |
| 0.4 | 2.0 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.3 | 0.9 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.3 | 12.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 1.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 1.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.3 | 5.0 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.3 | 1.8 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.3 | 3.8 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.2 | 25.1 | GO:0030017 | sarcomere(GO:0030017) |
| 0.2 | 2.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 25.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 4.7 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 6.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 7.2 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 2.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 3.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.4 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 2.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 4.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.9 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 3.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 25.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 8.0 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 1.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 4.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 0.8 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 0.7 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 0.7 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.7 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 2.0 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 1.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 3.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.4 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 2.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.9 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.6 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 1.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 2.6 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 3.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.7 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 17.6 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 0.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.2 | GO:0005521 | lamin binding(GO:0005521) |
| 1.6 | 4.7 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 1.5 | 4.6 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 1.5 | 19.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 1.4 | 8.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.1 | 12.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 1.1 | 4.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 1.0 | 3.0 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.8 | 2.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.8 | 2.3 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.7 | 2.2 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.7 | 7.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.7 | 2.0 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.6 | 5.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.5 | 2.8 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.4 | 1.7 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.4 | 2.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.4 | 2.8 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.4 | 1.6 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.4 | 1.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.4 | 1.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 1.4 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.3 | 5.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.3 | 10.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.3 | 1.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.3 | 1.9 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.3 | 2.4 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.2 | 2.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 4.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.2 | 1.8 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.2 | 2.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 0.8 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.2 | 0.9 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 5.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 3.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 1.9 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.2 | 3.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 1.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.9 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 0.7 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.1 | 0.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 2.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.3 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 0.3 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.1 | 1.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 3.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.8 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 7.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 25.7 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 1.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 3.6 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 1.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.9 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.7 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 1.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.4 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.1 | 1.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.6 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 1.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.4 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 1.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 2.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.9 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.6 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 1.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 5.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.7 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 2.1 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 2.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 3.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 6.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 3.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 1.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 6.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.9 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.6 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 0.4 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 1.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 3.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.0 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.8 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 1.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 6.4 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 12.8 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.8 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 5.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 38.7 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.5 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 7.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.1 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 1.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.3 | 3.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 3.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.3 | 3.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 3.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 1.7 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 2.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 6.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 3.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 0.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 5.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 9.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 5.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.8 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.6 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.2 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.1 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 19.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.7 | 4.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 2.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.3 | 6.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 3.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 1.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 4.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 1.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.2 | 1.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 6.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.8 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 1.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 5.2 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 1.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 0.9 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 0.9 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.6 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.1 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 1.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 2.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |