Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
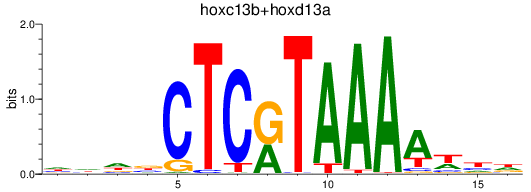
Results for hoxc13b+hoxd13a
Z-value: 1.18
Transcription factors associated with hoxc13b+hoxd13a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd13a
|
ENSDARG00000059256 | homeobox D13a |
|
hoxc13b
|
ENSDARG00000113877 | homeobox C13b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc13b | dr11_v1_chr11_+_2156430_2156430 | -0.59 | 5.8e-10 | Click! |
| hoxd13a | dr11_v1_chr9_-_1990323_1990323 | 0.18 | 8.8e-02 | Click! |
Activity profile of hoxc13b+hoxd13a motif
Sorted Z-values of hoxc13b+hoxd13a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_22637515 | 14.19 |
ENSDART00000158698
|
si:dkey-112a7.5
|
si:dkey-112a7.5 |
| chr22_-_24297510 | 13.22 |
ENSDART00000163297
|
si:ch211-117l17.6
|
si:ch211-117l17.6 |
| chr12_+_13256415 | 12.55 |
ENSDART00000144542
|
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr17_-_8886735 | 12.36 |
ENSDART00000121997
|
nkl.3
|
NK-lysin tandem duplicate 3 |
| chr17_+_2549503 | 12.14 |
ENSDART00000156843
|
si:dkey-248g15.3
|
si:dkey-248g15.3 |
| chr20_+_26683933 | 11.53 |
ENSDART00000139852
ENSDART00000077751 |
foxq1b
|
forkhead box Q1b |
| chr17_+_42274825 | 11.02 |
ENSDART00000020156
|
pax1a
|
paired box 1a |
| chr1_+_54650048 | 10.59 |
ENSDART00000141207
|
si:ch211-202h22.7
|
si:ch211-202h22.7 |
| chr9_-_33081978 | 10.33 |
ENSDART00000100918
|
zgc:172053
|
zgc:172053 |
| chr3_-_43872889 | 10.08 |
ENSDART00000170553
|
mslna
|
mesothelin a |
| chr1_-_59243542 | 9.61 |
ENSDART00000163021
|
mvb12a
|
multivesicular body subunit 12A |
| chr9_-_33063083 | 9.55 |
ENSDART00000048550
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr2_-_48196092 | 9.50 |
ENSDART00000139944
|
snorc
|
secondary ossification center associated regulator of chondrocyte maturation |
| chr1_+_45056371 | 8.57 |
ENSDART00000073689
ENSDART00000167309 |
btr01
|
bloodthirsty-related gene family, member 1 |
| chr17_-_37395460 | 8.56 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr2_+_37295088 | 8.53 |
ENSDART00000056519
|
gpr160
|
G protein-coupled receptor 160 |
| chr18_-_48547564 | 8.44 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr24_-_40668208 | 8.39 |
ENSDART00000171543
|
smyhc1
|
slow myosin heavy chain 1 |
| chr4_-_77551860 | 8.28 |
ENSDART00000188176
|
AL935186.6
|
|
| chr20_-_34127415 | 8.23 |
ENSDART00000010028
|
ptgs2b
|
prostaglandin-endoperoxide synthase 2b |
| chr10_+_35347993 | 8.17 |
ENSDART00000131350
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr25_+_29160102 | 8.14 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr14_+_10450216 | 8.11 |
ENSDART00000134814
|
cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr6_-_8360918 | 7.92 |
ENSDART00000004716
|
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr9_-_33081781 | 7.90 |
ENSDART00000165748
|
zgc:172053
|
zgc:172053 |
| chr25_-_3867990 | 7.84 |
ENSDART00000075663
|
cracr2b
|
calcium release activated channel regulator 2B |
| chr9_-_23990416 | 7.79 |
ENSDART00000113176
|
col6a3
|
collagen, type VI, alpha 3 |
| chr24_-_40667800 | 7.68 |
ENSDART00000169315
|
smyhc1
|
slow myosin heavy chain 1 |
| chr19_+_20778011 | 7.49 |
ENSDART00000024208
|
nutf2l
|
nuclear transport factor 2, like |
| chr15_-_35960250 | 7.23 |
ENSDART00000186765
|
col4a4
|
collagen, type IV, alpha 4 |
| chr21_-_35419486 | 7.12 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr4_+_55758103 | 7.03 |
ENSDART00000185964
|
CT583728.23
|
|
| chr18_+_48428713 | 6.97 |
ENSDART00000076861
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr13_-_34862452 | 6.88 |
ENSDART00000134573
ENSDART00000047552 |
sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr12_-_30523216 | 6.58 |
ENSDART00000152896
ENSDART00000153191 |
plekhs1
|
pleckstrin homology domain containing, family S member 1 |
| chr1_+_58282449 | 6.58 |
ENSDART00000131475
|
si:dkey-222h21.7
|
si:dkey-222h21.7 |
| chr20_-_38758797 | 6.58 |
ENSDART00000061394
|
trim54
|
tripartite motif containing 54 |
| chr22_+_24645325 | 6.50 |
ENSDART00000159531
|
lpar3
|
lysophosphatidic acid receptor 3 |
| chr9_-_9992697 | 6.44 |
ENSDART00000123415
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr1_-_681116 | 6.43 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr25_+_34014523 | 6.23 |
ENSDART00000182856
|
anxa2a
|
annexin A2a |
| chr16_-_12060770 | 6.22 |
ENSDART00000183237
ENSDART00000103948 |
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr9_-_33062891 | 6.22 |
ENSDART00000161182
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr18_+_2228737 | 6.16 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr16_+_50969248 | 5.97 |
ENSDART00000172068
|
si:dkeyp-97a10.2
|
si:dkeyp-97a10.2 |
| chr25_+_31277415 | 5.92 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr16_+_33144306 | 5.84 |
ENSDART00000101953
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr9_+_23770666 | 5.76 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr6_+_49771626 | 5.69 |
ENSDART00000134207
|
ctsz
|
cathepsin Z |
| chr6_+_49771372 | 5.68 |
ENSDART00000063251
|
ctsz
|
cathepsin Z |
| chr24_+_21621654 | 5.55 |
ENSDART00000002595
|
rpl21
|
ribosomal protein L21 |
| chr4_-_20118468 | 5.53 |
ENSDART00000078587
|
si:dkey-159a18.1
|
si:dkey-159a18.1 |
| chr16_+_33143503 | 5.33 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr5_+_37087583 | 5.16 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr25_+_31276842 | 5.11 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr24_-_40774028 | 5.09 |
ENSDART00000166578
|
CU633479.2
|
|
| chr16_+_33144112 | 5.04 |
ENSDART00000183149
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr5_+_57480014 | 5.02 |
ENSDART00000135520
|
si:ch211-202f5.3
|
si:ch211-202f5.3 |
| chr14_+_2487672 | 5.00 |
ENSDART00000170629
ENSDART00000123063 |
fgf18a
|
fibroblast growth factor 18a |
| chr10_+_26612321 | 4.98 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr6_-_39764995 | 4.95 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr10_+_24690534 | 4.89 |
ENSDART00000079549
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr3_-_25268751 | 4.88 |
ENSDART00000139423
|
mgat3a
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase a |
| chr4_-_74028161 | 4.87 |
ENSDART00000174062
|
LO018205.1
|
|
| chr16_-_12060488 | 4.86 |
ENSDART00000188733
|
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr16_+_35594670 | 4.75 |
ENSDART00000163275
|
si:ch211-1i11.3
|
si:ch211-1i11.3 |
| chr9_-_42730672 | 4.64 |
ENSDART00000136728
|
fkbp7
|
FK506 binding protein 7 |
| chr17_+_20607487 | 4.64 |
ENSDART00000154123
|
si:ch73-288o11.5
|
si:ch73-288o11.5 |
| chr24_-_25166416 | 4.64 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr18_-_35459605 | 4.56 |
ENSDART00000135691
|
itpkcb
|
inositol-trisphosphate 3-kinase Cb |
| chr16_-_36099492 | 4.52 |
ENSDART00000180905
|
CU499336.2
|
|
| chr7_+_22688781 | 4.42 |
ENSDART00000173509
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr16_-_21038015 | 4.40 |
ENSDART00000059239
|
snx10b
|
sorting nexin 10b |
| chr4_-_26413391 | 4.39 |
ENSDART00000145955
|
b4galnt3a
|
beta-1,4-N-acetyl-galactosaminyl transferase 3a |
| chr9_-_1965727 | 4.37 |
ENSDART00000082354
|
hoxd9a
|
homeobox D9a |
| chr5_-_67365750 | 4.30 |
ENSDART00000062359
|
unga
|
uracil DNA glycosylase a |
| chr2_-_36914281 | 4.25 |
ENSDART00000008322
|
si:dkey-193b15.5
|
si:dkey-193b15.5 |
| chr21_+_11503212 | 4.16 |
ENSDART00000146701
|
si:dkey-184p9.7
|
si:dkey-184p9.7 |
| chr2_+_48282590 | 4.14 |
ENSDART00000035338
|
lpar5a
|
lysophosphatidic acid receptor 5a |
| chr24_-_35561672 | 4.14 |
ENSDART00000058564
|
mcm4
|
minichromosome maintenance complex component 4 |
| chr4_-_30249792 | 4.05 |
ENSDART00000151833
ENSDART00000137564 ENSDART00000168249 |
si:dkey-265e15.2
|
si:dkey-265e15.2 |
| chr18_-_43866001 | 4.05 |
ENSDART00000150218
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr12_-_34435604 | 3.99 |
ENSDART00000115088
|
birc5a
|
baculoviral IAP repeat containing 5a |
| chr14_+_26247319 | 3.89 |
ENSDART00000192793
|
CCDC69
|
coiled-coil domain containing 69 |
| chr4_-_26095755 | 3.85 |
ENSDART00000100611
ENSDART00000191266 |
si:ch211-244b2.3
|
si:ch211-244b2.3 |
| chr19_-_25081711 | 3.84 |
ENSDART00000058513
|
xkr8.3
|
XK, Kell blood group complex subunit-related family, member 8, tandem duplicate 3 |
| chr7_+_17861489 | 3.81 |
ENSDART00000173860
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr8_-_12432604 | 3.80 |
ENSDART00000133350
ENSDART00000140699 ENSDART00000101174 |
traf1
|
TNF receptor-associated factor 1 |
| chr14_+_21699129 | 3.76 |
ENSDART00000073707
|
stx3a
|
syntaxin 3A |
| chr9_+_23714406 | 3.76 |
ENSDART00000189445
|
gypc
|
glycophorin C (Gerbich blood group) |
| chr20_+_23238833 | 3.74 |
ENSDART00000074167
|
ociad2
|
OCIA domain containing 2 |
| chr22_+_35205968 | 3.73 |
ENSDART00000150467
|
tsc22d2
|
TSC22 domain family 2 |
| chr6_+_48206535 | 3.67 |
ENSDART00000075172
|
cttnbp2nla
|
CTTNBP2 N-terminal like a |
| chr5_-_67365006 | 3.67 |
ENSDART00000136116
|
unga
|
uracil DNA glycosylase a |
| chr6_+_34028532 | 3.62 |
ENSDART00000155827
|
si:ch73-185c24.2
|
si:ch73-185c24.2 |
| chr22_+_18477934 | 3.62 |
ENSDART00000132684
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr15_-_41457898 | 3.54 |
ENSDART00000188847
|
nlrc9
|
NLR family CARD domain containing 9 |
| chr25_+_24616717 | 3.53 |
ENSDART00000089113
|
abtb2b
|
ankyrin repeat and BTB (POZ) domain containing 2b |
| chr12_+_17154655 | 3.52 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr20_+_18993452 | 3.47 |
ENSDART00000025509
|
tdh
|
L-threonine dehydrogenase |
| chr22_-_22164338 | 3.43 |
ENSDART00000183840
|
cdc34a
|
cell division cycle 34 homolog (S. cerevisiae) a |
| chr2_-_36599044 | 3.38 |
ENSDART00000161829
|
tradv23.3.8
|
T-cell receptor alpha/delta variable 23.3.8 |
| chr23_-_4933508 | 3.38 |
ENSDART00000137578
ENSDART00000141196 |
ngfa
|
nerve growth factor a (beta polypeptide) |
| chr24_-_18919562 | 3.30 |
ENSDART00000144244
ENSDART00000106188 ENSDART00000182518 |
cpa6
|
carboxypeptidase A6 |
| chr25_-_3230187 | 3.26 |
ENSDART00000160545
|
cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr6_+_7123980 | 3.26 |
ENSDART00000179738
ENSDART00000151311 |
si:ch211-237c6.4
|
si:ch211-237c6.4 |
| chr20_-_2713120 | 3.26 |
ENSDART00000138753
ENSDART00000104606 |
rars2
|
arginyl-tRNA synthetase 2, mitochondrial (putative) |
| chr18_-_43866526 | 3.19 |
ENSDART00000111309
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr1_+_54626491 | 3.16 |
ENSDART00000136063
|
si:ch211-202h22.9
|
si:ch211-202h22.9 |
| chr5_-_41841675 | 3.15 |
ENSDART00000141683
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr5_-_69621227 | 3.13 |
ENSDART00000178543
|
aldh2.2
|
aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 2 |
| chr3_-_29506960 | 3.10 |
ENSDART00000141720
|
cyth4a
|
cytohesin 4a |
| chr3_-_57666996 | 3.10 |
ENSDART00000166855
|
timp2b
|
TIMP metallopeptidase inhibitor 2b |
| chr11_-_2478374 | 3.09 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr14_-_38872536 | 3.08 |
ENSDART00000184315
ENSDART00000127479 |
gsr
|
glutathione reductase |
| chr15_-_14193926 | 3.07 |
ENSDART00000162707
|
pnkp
|
polynucleotide kinase 3'-phosphatase |
| chr15_-_14194208 | 2.98 |
ENSDART00000188237
ENSDART00000183155 ENSDART00000165520 |
pnkp
|
polynucleotide kinase 3'-phosphatase |
| chr4_-_32458327 | 2.96 |
ENSDART00000182527
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr3_+_24618012 | 2.96 |
ENSDART00000111997
|
zgc:171506
|
zgc:171506 |
| chr22_+_9764731 | 2.95 |
ENSDART00000161441
|
BX664625.4
|
|
| chr11_-_1400507 | 2.95 |
ENSDART00000173029
ENSDART00000172953 ENSDART00000111140 |
rpl29
|
ribosomal protein L29 |
| chr19_-_23249822 | 2.94 |
ENSDART00000140665
|
grb10a
|
growth factor receptor-bound protein 10a |
| chr4_+_60430885 | 2.92 |
ENSDART00000184608
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr11_+_3254252 | 2.89 |
ENSDART00000123568
|
pmela
|
premelanosome protein a |
| chr19_+_19756425 | 2.82 |
ENSDART00000167606
|
hoxa3a
|
homeobox A3a |
| chr5_-_69620722 | 2.81 |
ENSDART00000097248
|
aldh2.2
|
aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 2 |
| chr7_-_18508815 | 2.80 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr4_+_42419522 | 2.80 |
ENSDART00000135737
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr16_+_52343905 | 2.79 |
ENSDART00000131051
|
ifnlr1
|
interferon lambda receptor 1 |
| chr7_+_30051880 | 2.65 |
ENSDART00000075609
|
pstpip1b
|
proline-serine-threonine phosphatase interacting protein 1b |
| chr16_-_16761164 | 2.65 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr14_-_46259523 | 2.63 |
ENSDART00000172890
|
si:ch211-113d11.8
|
si:ch211-113d11.8 |
| chr8_+_23861461 | 2.61 |
ENSDART00000037109
|
srpk1a
|
SRSF protein kinase 1a |
| chr14_+_26439227 | 2.59 |
ENSDART00000054183
|
gpr137
|
G protein-coupled receptor 137 |
| chr3_-_31019715 | 2.59 |
ENSDART00000156687
ENSDART00000156127 ENSDART00000153945 |
si:dkeyp-71f10.5
|
si:dkeyp-71f10.5 |
| chr15_-_41332865 | 2.59 |
ENSDART00000170014
|
si:dkey-121n8.7
|
si:dkey-121n8.7 |
| chr15_-_41523225 | 2.57 |
ENSDART00000141202
ENSDART00000099290 ENSDART00000170395 ENSDART00000099437 |
si:dkey-230i18.2
si:dkey-121n8.7
nlrc9
|
si:dkey-230i18.2 si:dkey-121n8.7 NLR family CARD domain containing 9 |
| chr25_+_4787431 | 2.55 |
ENSDART00000170640
|
myo5c
|
myosin VC |
| chr6_+_28051978 | 2.53 |
ENSDART00000143218
|
si:ch73-194h10.2
|
si:ch73-194h10.2 |
| chr4_-_31984150 | 2.53 |
ENSDART00000162521
|
si:dkey-72l17.5
|
si:dkey-72l17.5 |
| chr4_+_15944245 | 2.52 |
ENSDART00000134594
|
si:dkey-117n7.3
|
si:dkey-117n7.3 |
| chr4_-_54381232 | 2.47 |
ENSDART00000157990
ENSDART00000169502 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr7_+_24881680 | 2.46 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr22_+_10676981 | 2.46 |
ENSDART00000138016
|
hyal2b
|
hyaluronoglucosaminidase 2b |
| chr17_+_12942634 | 2.45 |
ENSDART00000016597
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr3_-_27065477 | 2.42 |
ENSDART00000185660
|
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr3_-_23474416 | 2.40 |
ENSDART00000161672
|
si:dkey-225f5.5
|
si:dkey-225f5.5 |
| chr23_+_43638982 | 2.40 |
ENSDART00000168646
|
slc10a7
|
solute carrier family 10, member 7 |
| chr11_+_36675599 | 2.40 |
ENSDART00000170464
|
si:ch211-11c3.11
|
si:ch211-11c3.11 |
| chr13_-_9598320 | 2.38 |
ENSDART00000184613
|
cpxm1a
|
carboxypeptidase X (M14 family), member 1a |
| chr7_-_18617187 | 2.35 |
ENSDART00000172419
|
si:ch211-119e14.1
|
si:ch211-119e14.1 |
| chr8_+_18044257 | 2.33 |
ENSDART00000029255
|
glis1b
|
GLIS family zinc finger 1b |
| chr9_+_23003208 | 2.33 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr6_+_6924637 | 2.32 |
ENSDART00000065551
ENSDART00000151393 |
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr23_+_36083529 | 2.32 |
ENSDART00000053295
ENSDART00000130260 |
hoxc10a
|
homeobox C10a |
| chr19_+_7424347 | 2.31 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr18_+_40993369 | 2.31 |
ENSDART00000141162
|
si:dkey-283j8.1
|
si:dkey-283j8.1 |
| chr24_+_5799764 | 2.31 |
ENSDART00000154482
|
si:ch211-157j23.2
|
si:ch211-157j23.2 |
| chr22_+_5971404 | 2.28 |
ENSDART00000122153
|
BX322587.1
|
|
| chr4_+_41835660 | 2.23 |
ENSDART00000171428
|
si:ch211-268b14.2
|
si:ch211-268b14.2 |
| chr9_-_1959917 | 2.21 |
ENSDART00000082359
|
hoxd3a
|
homeobox D3a |
| chr5_+_32206378 | 2.16 |
ENSDART00000126873
ENSDART00000051361 |
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr21_-_40835069 | 2.15 |
ENSDART00000004686
|
limk1b
|
LIM domain kinase 1b |
| chr4_+_55059525 | 2.10 |
ENSDART00000128531
|
si:dkey-19c16.12
|
si:dkey-19c16.12 |
| chr12_-_7157527 | 2.00 |
ENSDART00000152274
|
si:dkey-187i8.2
|
si:dkey-187i8.2 |
| chr10_+_4987766 | 1.98 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr3_+_31058464 | 1.92 |
ENSDART00000153381
|
si:dkey-66i24.7
|
si:dkey-66i24.7 |
| chr4_-_71210030 | 1.92 |
ENSDART00000186473
|
si:ch211-205a14.7
|
si:ch211-205a14.7 |
| chr4_+_70913655 | 1.92 |
ENSDART00000180803
ENSDART00000185776 ENSDART00000193564 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr13_-_37649595 | 1.91 |
ENSDART00000115354
|
si:dkey-188i13.10
|
si:dkey-188i13.10 |
| chr8_+_22277198 | 1.91 |
ENSDART00000005989
|
dffb
|
DNA fragmentation factor, beta polypeptide (caspase-activated DNase) |
| chr4_-_67980261 | 1.91 |
ENSDART00000182305
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr17_+_2727807 | 1.88 |
ENSDART00000178759
|
kcnk10b
|
potassium channel, subfamily K, member 10b |
| chr19_-_27550768 | 1.84 |
ENSDART00000142313
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr4_-_37693537 | 1.83 |
ENSDART00000167526
|
si:dkey-3p4.1
|
si:dkey-3p4.1 |
| chr25_-_3892686 | 1.83 |
ENSDART00000043172
|
tmem258
|
transmembrane protein 258 |
| chr5_-_24008997 | 1.83 |
ENSDART00000066645
|
eif1axa
|
eukaryotic translation initiation factor 1A, X-linked, a |
| chr6_-_39649504 | 1.81 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr1_+_230363 | 1.79 |
ENSDART00000153285
|
tfdp1b
|
transcription factor Dp-1, b |
| chr2_-_51644044 | 1.78 |
ENSDART00000157899
|
dad1
|
defender against cell death 1 |
| chr20_-_54259780 | 1.77 |
ENSDART00000172631
|
fkbp3
|
FK506 binding protein 3 |
| chr5_+_66132394 | 1.74 |
ENSDART00000073892
|
zgc:114041
|
zgc:114041 |
| chr2_+_43749221 | 1.72 |
ENSDART00000142804
|
arhgap12a
|
Rho GTPase activating protein 12a |
| chr14_-_38873095 | 1.71 |
ENSDART00000047050
ENSDART00000173285 ENSDART00000147521 |
gsr
|
glutathione reductase |
| chr21_-_21641194 | 1.71 |
ENSDART00000010282
|
cbr1l
|
carbonyl reductase 1-like |
| chr10_-_31015535 | 1.70 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr18_+_40993196 | 1.68 |
ENSDART00000115111
|
si:dkey-283j8.1
|
si:dkey-283j8.1 |
| chr7_+_69653981 | 1.68 |
ENSDART00000090165
|
frem1a
|
Fras1 related extracellular matrix 1a |
| chr13_-_1349922 | 1.68 |
ENSDART00000140970
|
si:ch73-52p7.1
|
si:ch73-52p7.1 |
| chr3_+_43102010 | 1.67 |
ENSDART00000162096
|
micall2a
|
mical-like 2a |
| chr20_+_14789305 | 1.65 |
ENSDART00000002463
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr4_-_71151245 | 1.64 |
ENSDART00000165415
|
si:dkey-193i10.4
|
si:dkey-193i10.4 |
| chr13_+_45431660 | 1.63 |
ENSDART00000099950
|
syf2
|
SYF2 pre-mRNA-splicing factor |
| chr7_+_4592867 | 1.62 |
ENSDART00000192614
|
si:dkey-83f18.15
|
si:dkey-83f18.15 |
| chr11_+_3254524 | 1.61 |
ENSDART00000159459
|
pmela
|
premelanosome protein a |
| chr10_+_6907715 | 1.60 |
ENSDART00000041068
|
slc38a9
|
solute carrier family 38, member 9 |
| chr17_-_10122204 | 1.59 |
ENSDART00000160751
|
BX088587.1
|
|
| chr7_-_3639171 | 1.56 |
ENSDART00000064280
|
si:ch211-282j17.1
|
si:ch211-282j17.1 |
| chr2_-_11504778 | 1.56 |
ENSDART00000186556
|
sdr16c5a
|
short chain dehydrogenase/reductase family 16C, member 5a |
| chr5_+_30520249 | 1.54 |
ENSDART00000013431
|
hmbsa
|
hydroxymethylbilane synthase a |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc13b+hoxd13a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.6 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 2.9 | 8.6 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 2.4 | 7.2 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 2.3 | 16.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 2.2 | 6.5 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 2.1 | 8.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.9 | 9.3 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 1.5 | 4.5 | GO:0097435 | fibril organization(GO:0097435) |
| 0.8 | 3.8 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.6 | 6.9 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.6 | 4.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.6 | 4.1 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.5 | 3.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.4 | 4.9 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.4 | 4.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.4 | 3.5 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.4 | 2.8 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.4 | 1.2 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.4 | 1.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.3 | 3.8 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.3 | 1.0 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.3 | 2.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.3 | 1.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.3 | 4.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.3 | 4.9 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.3 | 3.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.3 | 1.1 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.3 | 1.9 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.3 | 1.3 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.3 | 1.5 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.2 | 5.9 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.2 | 0.7 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.2 | 1.7 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.2 | 0.7 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.2 | 1.2 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.2 | 3.0 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 1.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.2 | 4.6 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.2 | 2.9 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.2 | 0.8 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 | 7.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 3.8 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.2 | 4.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.2 | 14.2 | GO:0046939 | nucleotide phosphorylation(GO:0046939) |
| 0.2 | 1.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 | 1.0 | GO:1902914 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 3.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.2 | 7.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 6.2 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.2 | 7.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 0.4 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.1 | 0.7 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 3.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 2.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.8 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 2.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 2.2 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 2.2 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 7.9 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.1 | 0.8 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 2.3 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.1 | 0.7 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.1 | 0.7 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 2.1 | GO:0008406 | gonad development(GO:0008406) |
| 0.1 | 4.9 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 7.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 4.0 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.1 | 0.8 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 0.7 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 1.0 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 1.9 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 8.6 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.1 | 1.5 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 1.0 | GO:0035924 | cellular response to vascular endothelial growth factor stimulus(GO:0035924) |
| 0.1 | 8.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 9.3 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 4.5 | GO:0042493 | response to drug(GO:0042493) |
| 0.1 | 0.7 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.9 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.1 | 6.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 8.5 | GO:0001503 | ossification(GO:0001503) |
| 0.1 | 1.5 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.1 | 1.3 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 2.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 5.0 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 8.7 | GO:0051216 | cartilage development(GO:0051216) |
| 0.1 | 2.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 1.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 1.7 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 5.0 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.4 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.6 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 3.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.3 | GO:0019915 | lipid storage(GO:0019915) |
| 0.0 | 1.3 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 1.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.2 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 1.7 | GO:0033334 | fin morphogenesis(GO:0033334) |
| 0.0 | 2.8 | GO:0048884 | neuromast development(GO:0048884) |
| 0.0 | 1.4 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.1 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 1.0 | GO:0016441 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.0 | 1.3 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 2.7 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.8 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 5.3 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 7.3 | GO:0001525 | angiogenesis(GO:0001525) |
| 0.0 | 1.7 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:0045761 | cAMP biosynthetic process(GO:0006171) regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 1.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.8 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 3.2 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 2.9 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.2 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 5.4 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.1 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 2.0 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 3.0 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.0 | 1.1 | GO:0006518 | peptide metabolic process(GO:0006518) |
| 0.0 | 1.1 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.9 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.7 | 6.9 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.5 | 1.6 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.5 | 12.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.4 | 4.9 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.3 | 5.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.3 | 2.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.3 | 2.4 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.3 | 3.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 11.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 4.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 1.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.2 | 26.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 7.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 1.3 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.2 | 0.7 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 15.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.9 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 8.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.4 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.1 | 0.8 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 4.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 2.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 3.1 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 6.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 3.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 4.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 6.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 12.7 | GO:0000323 | lytic vacuole(GO:0000323) |
| 0.0 | 11.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.1 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 1.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 15.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.5 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 2.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 4.5 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.2 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 2.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 4.3 | GO:0043235 | receptor complex(GO:0043235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.3 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 2.4 | 7.2 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 2.1 | 8.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 2.0 | 6.1 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 1.4 | 8.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.2 | 8.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 1.0 | 7.9 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 1.0 | 6.9 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.9 | 4.4 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.8 | 4.9 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.8 | 3.0 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.7 | 12.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.7 | 3.5 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.6 | 5.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.6 | 3.0 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.5 | 6.2 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.5 | 6.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.4 | 4.9 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.4 | 1.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.4 | 2.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.4 | 5.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 4.0 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.3 | 0.9 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.3 | 4.6 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.3 | 3.4 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.3 | 1.5 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.2 | 2.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 3.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 3.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 5.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 0.6 | GO:0000035 | acyl binding(GO:0000035) |
| 0.2 | 4.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 1.6 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.2 | 0.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.2 | 10.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 1.0 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 0.7 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.2 | 4.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 9.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.2 | 8.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 6.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 3.3 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 0.6 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.4 | GO:0045547 | dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.1 | 3.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.8 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.7 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 1.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 18.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 1.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.6 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.8 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 3.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 16.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 7.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.8 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 1.7 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 1.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 4.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 7.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 9.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 1.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 1.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.0 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 1.0 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 1.7 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 5.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 1.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 7.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 2.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 10.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 2.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.5 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 2.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.4 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 3.8 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 1.1 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 1.1 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 22.5 | GO:0038023 | signaling receptor activity(GO:0038023) |
| 0.0 | 2.8 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 3.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 1.3 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 3.3 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.7 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 1.0 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 2.0 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 15.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 2.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 5.7 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.2 | 4.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 8.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 16.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.9 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 4.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 1.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 4.2 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 1.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 7.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.8 | 8.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.4 | 11.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.4 | 1.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.4 | 13.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 4.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 4.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 3.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 3.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 8.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 6.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 0.8 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 1.6 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 1.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.7 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.8 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |