Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
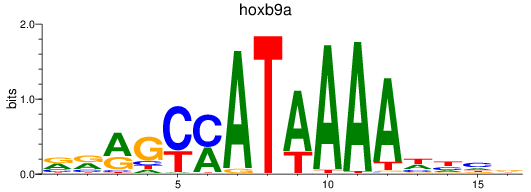
Results for hoxb9a
Z-value: 0.52
Transcription factors associated with hoxb9a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb9a
|
ENSDARG00000056023 | homeobox B9a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb9a | dr11_v1_chr3_+_23677351_23677351 | -0.14 | 1.9e-01 | Click! |
Activity profile of hoxb9a motif
Sorted Z-values of hoxb9a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_7379728 | 9.40 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr12_-_20373058 | 7.00 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr4_+_7391110 | 4.28 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr11_-_18253111 | 4.07 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr2_+_52847049 | 3.89 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr15_+_20403903 | 3.86 |
ENSDART00000134182
|
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr1_+_17900306 | 3.56 |
ENSDART00000089480
|
cyp4v8
|
cytochrome P450, family 4, subfamily V, polypeptide 8 |
| chr9_-_9980704 | 3.34 |
ENSDART00000130243
ENSDART00000193475 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr23_+_10352921 | 3.10 |
ENSDART00000081193
|
KRT18 (1 of many)
|
si:ch211-133j6.3 |
| chr11_+_24314148 | 3.09 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr23_+_18722915 | 3.06 |
ENSDART00000025057
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr23_+_18722715 | 3.05 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr24_+_39105051 | 2.94 |
ENSDART00000115297
|
mss51
|
MSS51 mitochondrial translational activator |
| chr23_+_39606108 | 2.80 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr1_-_9249943 | 2.73 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr21_+_11969603 | 2.57 |
ENSDART00000142247
ENSDART00000140652 |
mlnl
|
motilin-like |
| chr21_-_19918286 | 2.29 |
ENSDART00000180816
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr15_+_45586471 | 2.26 |
ENSDART00000165699
|
cldn15lb
|
claudin 15-like b |
| chr20_+_52546186 | 1.80 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr11_+_44503774 | 1.76 |
ENSDART00000169295
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr21_-_2415808 | 1.72 |
ENSDART00000171179
|
si:ch211-241b2.5
|
si:ch211-241b2.5 |
| chr18_-_48296793 | 1.69 |
ENSDART00000032184
ENSDART00000193076 |
CABZ01069595.1
|
|
| chr12_-_33817114 | 1.62 |
ENSDART00000161265
|
twnk
|
twinkle mtDNA helicase |
| chr25_+_32755485 | 1.60 |
ENSDART00000162188
|
etfa
|
electron-transfer-flavoprotein, alpha polypeptide |
| chr25_+_29161609 | 1.53 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr10_-_7785930 | 1.53 |
ENSDART00000043961
ENSDART00000111058 |
mpx
|
myeloid-specific peroxidase |
| chr24_-_32025637 | 1.53 |
ENSDART00000180448
ENSDART00000159034 |
rsu1
|
Ras suppressor protein 1 |
| chr14_+_15495088 | 1.52 |
ENSDART00000165765
ENSDART00000188577 |
si:dkey-203a12.6
|
si:dkey-203a12.6 |
| chr7_+_27317174 | 1.49 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr11_-_25538341 | 1.46 |
ENSDART00000171560
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr24_+_1042594 | 1.42 |
ENSDART00000109117
|
si:dkey-192l18.9
|
si:dkey-192l18.9 |
| chr8_-_38159805 | 1.31 |
ENSDART00000112331
ENSDART00000180006 |
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr15_+_32405959 | 1.31 |
ENSDART00000177269
|
si:ch211-162k9.6
|
si:ch211-162k9.6 |
| chr10_+_44700103 | 1.29 |
ENSDART00000165999
|
scarb1
|
scavenger receptor class B, member 1 |
| chr17_+_33375469 | 1.27 |
ENSDART00000032827
|
zgc:162964
|
zgc:162964 |
| chr17_+_15788100 | 1.26 |
ENSDART00000027667
|
rragd
|
ras-related GTP binding D |
| chr10_+_40321067 | 1.22 |
ENSDART00000109816
|
gltpb
|
glycolipid transfer protein b |
| chr3_+_36646054 | 1.15 |
ENSDART00000170013
ENSDART00000159948 |
gspt1l
|
G1 to S phase transition 1, like |
| chr5_-_32336613 | 1.14 |
ENSDART00000139732
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr20_-_2641233 | 1.14 |
ENSDART00000145335
ENSDART00000133121 |
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr9_-_22355391 | 1.11 |
ENSDART00000009115
|
crygm3
|
crystallin, gamma M3 |
| chr7_+_24522308 | 1.11 |
ENSDART00000173542
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr24_-_9997948 | 1.11 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr15_-_21014015 | 1.10 |
ENSDART00000144991
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr10_+_15107886 | 1.09 |
ENSDART00000188047
ENSDART00000164095 |
scpp8
|
secretory calcium-binding phosphoprotein 8 |
| chr7_+_33152723 | 1.08 |
ENSDART00000132658
|
si:ch211-194p6.10
|
si:ch211-194p6.10 |
| chr11_-_15090118 | 1.08 |
ENSDART00000171118
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr24_-_14712427 | 1.07 |
ENSDART00000176316
|
jph1a
|
junctophilin 1a |
| chr1_+_44439661 | 1.07 |
ENSDART00000100309
|
crybb1l2
|
crystallin, beta B1, like 2 |
| chr5_+_72194444 | 1.02 |
ENSDART00000165436
|
ddx54
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 |
| chr3_-_49554912 | 0.99 |
ENSDART00000159392
|
CR847534.1
|
|
| chr15_+_29727799 | 0.99 |
ENSDART00000182006
|
zgc:153372
|
zgc:153372 |
| chr2_+_15586632 | 0.98 |
ENSDART00000164903
|
BX510342.1
|
|
| chr16_-_27566552 | 0.97 |
ENSDART00000142102
|
zgc:153215
|
zgc:153215 |
| chr13_-_33362191 | 0.97 |
ENSDART00000100514
|
zgc:172120
|
zgc:172120 |
| chr13_-_11578304 | 0.96 |
ENSDART00000171145
|
si:ch211-132e22.4
|
si:ch211-132e22.4 |
| chr8_+_45004997 | 0.96 |
ENSDART00000159779
|
si:ch211-163b2.4
|
si:ch211-163b2.4 |
| chr8_+_45004666 | 0.93 |
ENSDART00000145348
|
si:ch211-163b2.4
|
si:ch211-163b2.4 |
| chr7_-_7692723 | 0.92 |
ENSDART00000183352
|
aadat
|
aminoadipate aminotransferase |
| chr10_+_40660772 | 0.92 |
ENSDART00000148007
|
taar19l
|
trace amine associated receptor 19l |
| chr10_+_26612321 | 0.92 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr3_-_34089310 | 0.91 |
ENSDART00000151774
|
ighv5-5
|
immunoglobulin heavy variable 5-5 |
| chr17_-_43666166 | 0.91 |
ENSDART00000077990
|
egr2a
|
early growth response 2a |
| chr3_-_34624745 | 0.91 |
ENSDART00000151091
|
tac4
|
tachykinin 4 (hemokinin) |
| chr21_-_39931285 | 0.90 |
ENSDART00000180010
ENSDART00000024407 |
tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr7_-_18508815 | 0.90 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr23_-_21215311 | 0.87 |
ENSDART00000112424
|
megf6a
|
multiple EGF-like-domains 6a |
| chr5_-_57723929 | 0.87 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr7_-_7692992 | 0.86 |
ENSDART00000192619
|
aadat
|
aminoadipate aminotransferase |
| chr16_-_16225260 | 0.86 |
ENSDART00000165790
|
gra
|
granulito |
| chr21_-_39024754 | 0.86 |
ENSDART00000056878
|
traf4b
|
tnf receptor-associated factor 4b |
| chr5_-_9625459 | 0.85 |
ENSDART00000143347
|
sh2b3
|
SH2B adaptor protein 3 |
| chr2_-_40196547 | 0.85 |
ENSDART00000168098
|
ccl34a.3
|
chemokine (C-C motif) ligand 34a, duplicate 3 |
| chr12_+_27704015 | 0.84 |
ENSDART00000153256
|
cacna1g
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr6_+_13083146 | 0.84 |
ENSDART00000172158
|
l3hypdh
|
trans-L-3-hydroxyproline dehydratase |
| chr10_+_506538 | 0.83 |
ENSDART00000141713
|
si:ch211-242f23.3
|
si:ch211-242f23.3 |
| chr21_-_11970199 | 0.82 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr15_-_2841677 | 0.82 |
ENSDART00000026145
ENSDART00000180290 |
AMOTL1
|
angiomotin like 1 |
| chr3_+_58119571 | 0.81 |
ENSDART00000108979
|
si:ch211-256e16.4
|
si:ch211-256e16.4 |
| chr22_-_4644484 | 0.80 |
ENSDART00000167748
|
fbn2b
|
fibrillin 2b |
| chr4_-_77300601 | 0.79 |
ENSDART00000163015
ENSDART00000174015 |
slco1f2
|
solute carrier organic anion transporter family, member 1F2 |
| chr2_-_3158919 | 0.78 |
ENSDART00000098394
|
wnt3a
|
wingless-type MMTV integration site family, member 3A |
| chr11_-_15090564 | 0.75 |
ENSDART00000162079
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr24_-_31452875 | 0.75 |
ENSDART00000187381
ENSDART00000185128 |
cngb3.2
|
cyclic nucleotide gated channel beta 3, tandem duplicate 2 |
| chr10_-_40484531 | 0.74 |
ENSDART00000142464
ENSDART00000180318 |
taar20z
|
trace amine associated receptor 20z |
| chr19_+_19747430 | 0.71 |
ENSDART00000166129
|
hoxa9a
|
homeobox A9a |
| chr17_-_30652738 | 0.71 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr11_-_38533505 | 0.71 |
ENSDART00000113894
|
slc45a3
|
solute carrier family 45, member 3 |
| chr23_+_3625083 | 0.71 |
ENSDART00000184958
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr10_+_40768203 | 0.70 |
ENSDART00000171994
ENSDART00000140343 |
taar19d
|
trace amine associated receptor 19d |
| chr25_-_19219807 | 0.70 |
ENSDART00000183577
|
acanb
|
aggrecan b |
| chr5_+_24087035 | 0.69 |
ENSDART00000183644
|
tp53
|
tumor protein p53 |
| chr15_+_31515976 | 0.68 |
ENSDART00000156471
|
tex26
|
testis expressed 26 |
| chr13_+_1182257 | 0.68 |
ENSDART00000033528
ENSDART00000183702 ENSDART00000147959 |
tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr20_+_46202188 | 0.67 |
ENSDART00000100523
|
taar13c
|
trace amine associated receptor 13c |
| chr2_+_19578079 | 0.67 |
ENSDART00000144413
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr1_+_56463494 | 0.66 |
ENSDART00000097964
|
zgc:171452
|
zgc:171452 |
| chr23_+_36122058 | 0.66 |
ENSDART00000184448
|
hoxc3a
|
homeobox C3a |
| chr17_-_36841981 | 0.66 |
ENSDART00000131566
|
myo6b
|
myosin VIb |
| chr24_-_3426620 | 0.65 |
ENSDART00000184346
|
nck1b
|
NCK adaptor protein 1b |
| chr20_-_2713120 | 0.65 |
ENSDART00000138753
ENSDART00000104606 |
rars2
|
arginyl-tRNA synthetase 2, mitochondrial (putative) |
| chr25_-_31898552 | 0.65 |
ENSDART00000156128
|
si:ch73-330k17.3
|
si:ch73-330k17.3 |
| chr24_+_30392834 | 0.64 |
ENSDART00000162555
|
dpyda.1
|
dihydropyrimidine dehydrogenase a, tandem duplicate 1 |
| chr4_-_11122638 | 0.64 |
ENSDART00000150600
|
si:dkey-21h14.10
|
si:dkey-21h14.10 |
| chr7_+_29512673 | 0.63 |
ENSDART00000173895
|
si:dkey-182o15.5
|
si:dkey-182o15.5 |
| chr2_-_32637592 | 0.62 |
ENSDART00000136353
|
si:dkeyp-73d8.8
|
si:dkeyp-73d8.8 |
| chr10_-_3332362 | 0.62 |
ENSDART00000007577
ENSDART00000055140 |
tor4aa
|
torsin family 4, member Aa |
| chr11_+_12175162 | 0.60 |
ENSDART00000125446
|
si:ch211-156l18.7
|
si:ch211-156l18.7 |
| chr11_+_42585138 | 0.58 |
ENSDART00000019008
|
asb14a
|
ankyrin repeat and SOCS box containing 14a |
| chr14_+_22447662 | 0.58 |
ENSDART00000161776
|
sowahab
|
sosondowah ankyrin repeat domain family member Ab |
| chr11_+_45323455 | 0.57 |
ENSDART00000189249
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr8_-_6897619 | 0.57 |
ENSDART00000133606
|
si:ch211-255g12.8
|
si:ch211-255g12.8 |
| chr20_-_31496679 | 0.56 |
ENSDART00000153437
ENSDART00000153193 |
sash1a
|
SAM and SH3 domain containing 1a |
| chr4_+_59711338 | 0.53 |
ENSDART00000150849
|
si:dkey-149m13.4
|
si:dkey-149m13.4 |
| chr6_-_54107269 | 0.52 |
ENSDART00000190017
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr4_-_64142389 | 0.51 |
ENSDART00000172126
|
BX914205.3
|
|
| chr10_-_1625080 | 0.51 |
ENSDART00000137285
|
nup155
|
nucleoporin 155 |
| chr4_+_45504938 | 0.51 |
ENSDART00000145958
|
si:dkey-256i11.2
|
si:dkey-256i11.2 |
| chr20_-_47270519 | 0.48 |
ENSDART00000153329
|
si:dkeyp-104f11.8
|
si:dkeyp-104f11.8 |
| chr5_-_39698224 | 0.48 |
ENSDART00000076929
|
prkg2
|
protein kinase, cGMP-dependent, type II |
| chr17_-_50050453 | 0.47 |
ENSDART00000182057
|
zgc:100951
|
zgc:100951 |
| chr13_+_27329556 | 0.47 |
ENSDART00000140085
|
mb21d1
|
Mab-21 domain containing 1 |
| chr11_-_38492269 | 0.47 |
ENSDART00000065613
|
elk4
|
ELK4, ETS-domain protein |
| chr17_-_21057617 | 0.46 |
ENSDART00000148095
ENSDART00000048853 |
ube2d1a
|
ubiquitin-conjugating enzyme E2D 1a |
| chr5_-_30074332 | 0.44 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr4_-_59159690 | 0.42 |
ENSDART00000164706
|
znf1149
|
zinc finger protein 1149 |
| chr24_-_21819010 | 0.42 |
ENSDART00000091096
|
CR352265.1
|
|
| chr22_+_8612462 | 0.39 |
ENSDART00000114586
|
CR450686.1
|
|
| chr17_-_23727978 | 0.39 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr18_+_8320165 | 0.39 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr4_+_33574463 | 0.38 |
ENSDART00000150255
|
si:dkey-84h14.1
|
si:dkey-84h14.1 |
| chr4_-_50788075 | 0.38 |
ENSDART00000150302
|
znf1045
|
zinc finger protein 1045 |
| chr10_+_44699734 | 0.37 |
ENSDART00000167952
ENSDART00000158681 ENSDART00000190188 ENSDART00000168276 |
scarb1
|
scavenger receptor class B, member 1 |
| chr4_-_61920018 | 0.36 |
ENSDART00000164832
|
znf1056
|
zinc finger protein 1056 |
| chr17_-_50059664 | 0.35 |
ENSDART00000193940
|
FO704836.1
|
|
| chr6_+_39536596 | 0.34 |
ENSDART00000185742
|
CU467856.3
|
|
| chr22_+_2512154 | 0.33 |
ENSDART00000097363
|
zgc:173726
|
zgc:173726 |
| chr22_+_6321038 | 0.32 |
ENSDART00000179961
|
si:rp71-1i20.1
|
si:rp71-1i20.1 |
| chr25_-_12923482 | 0.31 |
ENSDART00000161754
|
CR450808.1
|
|
| chr10_+_40700311 | 0.31 |
ENSDART00000157650
ENSDART00000138342 |
taar19n
|
trace amine associated receptor 19n |
| chr3_+_13482542 | 0.29 |
ENSDART00000161287
|
CABZ01079251.1
|
|
| chr5_-_54424019 | 0.29 |
ENSDART00000169270
ENSDART00000164067 |
coq4
|
coenzyme Q4 homolog (S. cerevisiae) |
| chr3_+_6469754 | 0.29 |
ENSDART00000185809
|
NUP85 (1 of many)
|
nucleoporin 85 |
| chr8_-_7847921 | 0.27 |
ENSDART00000188094
|
zgc:113363
|
zgc:113363 |
| chr22_+_21255860 | 0.26 |
ENSDART00000134893
|
cpamd8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr24_+_29352039 | 0.25 |
ENSDART00000101641
|
prmt6
|
protein arginine methyltransferase 6 |
| chr5_-_30984271 | 0.24 |
ENSDART00000051392
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr4_-_71108793 | 0.24 |
ENSDART00000189856
|
si:dkeyp-80d11.1
|
si:dkeyp-80d11.1 |
| chr10_+_16069987 | 0.23 |
ENSDART00000043936
|
megf10
|
multiple EGF-like-domains 10 |
| chr1_-_56556326 | 0.23 |
ENSDART00000188760
|
CABZ01059408.1
|
|
| chr21_-_27251112 | 0.22 |
ENSDART00000137667
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr4_-_55641422 | 0.22 |
ENSDART00000165178
|
znf1074
|
zinc finger protein 1074 |
| chr25_+_28823952 | 0.22 |
ENSDART00000067072
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr4_-_46915962 | 0.22 |
ENSDART00000169555
|
si:ch211-134c10.1
|
si:ch211-134c10.1 |
| chr6_-_6345991 | 0.18 |
ENSDART00000174445
|
ccdc88aa
|
coiled-coil domain containing 88Aa |
| chr6_-_3982783 | 0.17 |
ENSDART00000171944
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr25_+_7299488 | 0.17 |
ENSDART00000184836
|
hmg20a
|
high mobility group 20A |
| chr13_+_22295905 | 0.17 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr13_-_2283176 | 0.16 |
ENSDART00000158462
|
lrrc1
|
leucine rich repeat containing 1 |
| chr25_+_32496877 | 0.15 |
ENSDART00000132698
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr25_+_22572296 | 0.14 |
ENSDART00000152096
|
stra6
|
stimulated by retinoic acid 6 |
| chr7_+_31145386 | 0.14 |
ENSDART00000075407
ENSDART00000169462 |
fam189a1
|
family with sequence similarity 189, member A1 |
| chr3_-_53091946 | 0.14 |
ENSDART00000187297
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr6_+_19948043 | 0.14 |
ENSDART00000182636
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr8_-_3312384 | 0.14 |
ENSDART00000035965
|
fut9b
|
fucosyltransferase 9b |
| chr4_+_42556555 | 0.12 |
ENSDART00000168536
|
znf1053
|
zinc finger protein 1053 |
| chr21_+_23108420 | 0.12 |
ENSDART00000192394
ENSDART00000088459 |
htr3b
|
5-hydroxytryptamine (serotonin) receptor 3B |
| chr6_+_41186320 | 0.12 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr18_+_35173859 | 0.11 |
ENSDART00000127379
ENSDART00000098292 |
cfap45
|
cilia and flagella associated protein 45 |
| chr4_-_73190246 | 0.11 |
ENSDART00000170842
|
LO018260.1
|
|
| chr2_+_2967255 | 0.11 |
ENSDART00000167649
ENSDART00000166449 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr21_-_3700334 | 0.11 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr20_+_33532296 | 0.08 |
ENSDART00000153153
|
kcnf1a
|
potassium voltage-gated channel, subfamily F, member 1a |
| chr7_-_41693004 | 0.07 |
ENSDART00000121509
|
malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr3_-_25490970 | 0.05 |
ENSDART00000184651
|
grb2b
|
growth factor receptor-bound protein 2b |
| chr19_-_24218942 | 0.05 |
ENSDART00000189198
|
BX547993.2
|
|
| chr18_+_35173683 | 0.04 |
ENSDART00000192545
|
cfap45
|
cilia and flagella associated protein 45 |
| chr10_-_10969444 | 0.04 |
ENSDART00000138041
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr6_+_41956355 | 0.03 |
ENSDART00000176856
|
oxtr
|
oxytocin receptor |
| chr20_-_34670236 | 0.02 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr10_-_42923385 | 0.01 |
ENSDART00000076731
|
ACOT12
|
acyl-CoA thioesterase 12 |
| chr10_-_22127942 | 0.01 |
ENSDART00000133374
|
ponzr2
|
plac8 onzin related protein 2 |
| chr3_+_33745014 | 0.00 |
ENSDART00000159966
|
nacc1a
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing a |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb9a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.4 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 1.2 | 3.6 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.8 | 4.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.8 | 2.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.6 | 3.9 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.6 | 3.1 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.6 | 1.7 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.4 | 1.3 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.3 | 1.0 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.3 | 1.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 | 0.8 | GO:0060958 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.2 | 1.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 1.6 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.2 | 0.7 | GO:1901216 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.2 | 0.9 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 0.9 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 1.6 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.2 | 1.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 0.5 | GO:0090435 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.2 | 1.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.2 | 1.7 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 1.5 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 1.1 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.1 | 0.7 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.6 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.1 | 1.5 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.1 | 0.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 3.9 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 0.8 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 0.9 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 2.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.6 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 4.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.7 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.2 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.0 | 0.8 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.4 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 1.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.9 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 1.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 2.7 | GO:0034976 | response to endoplasmic reticulum stress(GO:0034976) |
| 0.0 | 0.7 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 2.6 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.5 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 1.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.8 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.7 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 2.2 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 1.0 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 1.4 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.3 | 1.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.3 | 1.3 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 1.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 1.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 2.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.5 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 4.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.1 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 6.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 3.9 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 1.7 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.9 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 7.0 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 1.7 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 1.0 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.3 | 0.9 | GO:0031835 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.3 | 1.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 1.8 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.2 | 1.2 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.2 | 3.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 2.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 3.9 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.8 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.5 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.6 | GO:0017113 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.1 | 0.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.8 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 3.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.9 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 1.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.4 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.1 | 0.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.7 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 1.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 3.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.7 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.0 | 0.4 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 1.6 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 1.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 2.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 2.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 6.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 5.1 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.7 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.6 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 1.0 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.8 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 10.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.5 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 1.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |