Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
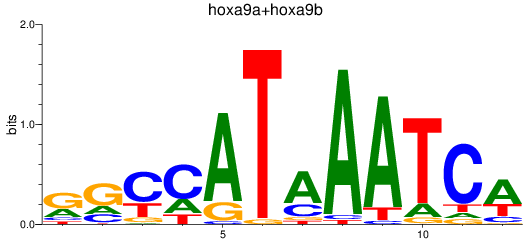
Results for hoxa9a+hoxa9b
Z-value: 1.39
Transcription factors associated with hoxa9a+hoxa9b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa9b
|
ENSDARG00000056819 | homeobox A9b |
|
hoxa9a
|
ENSDARG00000105013 | homeobox A9a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa9b | dr11_v1_chr16_+_20915319_20915319 | 0.46 | 2.6e-06 | Click! |
| hoxa9a | dr11_v1_chr19_+_19750101_19750223 | 0.31 | 2.7e-03 | Click! |
Activity profile of hoxa9a+hoxa9b motif
Sorted Z-values of hoxa9a+hoxa9b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_+_38301080 | 38.21 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr7_+_39386982 | 22.52 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr3_+_32526263 | 21.54 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr5_-_32274383 | 19.41 |
ENSDART00000122889
|
myhz1.3
|
myosin, heavy polypeptide 1.3, skeletal muscle |
| chr8_+_6576940 | 16.80 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr6_-_46875310 | 16.64 |
ENSDART00000154442
|
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr6_+_55032439 | 16.52 |
ENSDART00000164232
ENSDART00000158845 ENSDART00000157584 ENSDART00000026359 ENSDART00000122794 ENSDART00000183742 |
mybphb
|
myosin binding protein Hb |
| chr3_+_33300522 | 15.83 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr19_-_10425140 | 15.10 |
ENSDART00000145319
|
si:ch211-171h4.3
|
si:ch211-171h4.3 |
| chr5_+_32206378 | 14.77 |
ENSDART00000126873
ENSDART00000051361 |
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr1_+_7546259 | 13.74 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr7_+_69841017 | 13.55 |
ENSDART00000169107
|
FO818704.1
|
|
| chr11_-_6048490 | 12.59 |
ENSDART00000066164
|
plvapb
|
plasmalemma vesicle associated protein b |
| chr15_+_32711172 | 11.29 |
ENSDART00000163936
ENSDART00000168135 |
postnb
|
periostin, osteoblast specific factor b |
| chr8_-_2616326 | 10.98 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr21_+_11969603 | 10.69 |
ENSDART00000142247
ENSDART00000140652 |
mlnl
|
motilin-like |
| chr5_+_2815021 | 10.69 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr1_-_681116 | 10.29 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr16_+_34111919 | 9.86 |
ENSDART00000134037
ENSDART00000006061 ENSDART00000140552 |
tcea3
|
transcription elongation factor A (SII), 3 |
| chr7_+_31879649 | 9.74 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr1_-_33647138 | 9.33 |
ENSDART00000142111
ENSDART00000015547 |
cldng
|
claudin g |
| chr3_+_46425262 | 9.25 |
ENSDART00000153690
|
sc:d0202
|
sc:d0202 |
| chr17_+_1323699 | 9.14 |
ENSDART00000172540
|
adssl1
|
adenylosuccinate synthase like 1 |
| chr7_+_31879986 | 8.99 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr20_+_6142433 | 8.58 |
ENSDART00000054084
ENSDART00000136986 |
ttr
|
transthyretin (prealbumin, amyloidosis type I) |
| chr7_-_69853453 | 8.38 |
ENSDART00000049928
|
myoz2a
|
myozenin 2a |
| chr16_-_13516745 | 8.16 |
ENSDART00000145410
|
si:dkeyp-69b9.3
|
si:dkeyp-69b9.3 |
| chr12_-_6172154 | 7.80 |
ENSDART00000185434
|
a1cf
|
apobec1 complementation factor |
| chr4_-_16354292 | 7.78 |
ENSDART00000139919
|
lum
|
lumican |
| chr25_-_3830272 | 7.75 |
ENSDART00000055843
|
cd151
|
CD151 molecule |
| chr9_-_9992697 | 7.75 |
ENSDART00000123415
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr19_+_17385561 | 7.74 |
ENSDART00000141397
ENSDART00000143913 ENSDART00000133626 |
rpl15
|
ribosomal protein L15 |
| chr11_+_36243774 | 7.57 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr8_-_23599096 | 7.30 |
ENSDART00000183096
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr13_+_13681681 | 7.30 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr16_+_12836143 | 7.28 |
ENSDART00000067741
|
cacng6b
|
calcium channel, voltage-dependent, gamma subunit 6b |
| chr7_-_4461104 | 7.23 |
ENSDART00000023090
ENSDART00000140770 |
slc12a10.1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 1 |
| chr20_-_33959815 | 7.22 |
ENSDART00000133081
|
selp
|
selectin P |
| chr2_-_48196092 | 7.20 |
ENSDART00000139944
|
snorc
|
secondary ossification center associated regulator of chondrocyte maturation |
| chr3_-_3209432 | 7.18 |
ENSDART00000140635
|
si:ch211-229i14.2
|
si:ch211-229i14.2 |
| chr12_-_26406323 | 7.10 |
ENSDART00000131896
|
myoz1b
|
myozenin 1b |
| chr5_-_16351306 | 7.05 |
ENSDART00000168643
|
CABZ01088700.1
|
|
| chr22_+_28446365 | 6.98 |
ENSDART00000189359
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr24_-_12938922 | 6.95 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr9_-_6380653 | 6.80 |
ENSDART00000078523
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr22_+_28446557 | 6.76 |
ENSDART00000089546
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr15_+_21202820 | 6.72 |
ENSDART00000154036
|
si:dkey-52d15.2
|
si:dkey-52d15.2 |
| chr7_+_26545911 | 6.60 |
ENSDART00000135313
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr4_-_16353733 | 6.58 |
ENSDART00000186785
|
lum
|
lumican |
| chr7_+_35268880 | 6.51 |
ENSDART00000182231
|
dpep2
|
dipeptidase 2 |
| chr7_+_26545502 | 6.32 |
ENSDART00000140528
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr16_+_1254390 | 6.32 |
ENSDART00000092627
|
ADAMTSL4
|
ADAMTS like 4 |
| chr18_-_46010 | 6.29 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr9_-_23807032 | 6.15 |
ENSDART00000027443
|
esyt3
|
extended synaptotagmin-like protein 3 |
| chr8_+_3405612 | 6.14 |
ENSDART00000163437
|
zgc:112433
|
zgc:112433 |
| chr21_+_18997511 | 6.11 |
ENSDART00000145591
|
rpl17
|
ribosomal protein L17 |
| chr6_+_50451337 | 6.10 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr20_-_463786 | 6.08 |
ENSDART00000032212
|
frk
|
fyn-related Src family tyrosine kinase |
| chr19_-_31042570 | 5.99 |
ENSDART00000144337
ENSDART00000136213 ENSDART00000133101 ENSDART00000190949 |
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr19_-_27550768 | 5.99 |
ENSDART00000142313
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr10_-_8053753 | 5.94 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr23_-_31506854 | 5.89 |
ENSDART00000131352
ENSDART00000138625 ENSDART00000133002 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr19_+_19775757 | 5.81 |
ENSDART00000164677
|
hoxa3a
|
homeobox A3a |
| chr20_-_38758797 | 5.79 |
ENSDART00000061394
|
trim54
|
tripartite motif containing 54 |
| chr14_-_36799280 | 5.74 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr25_+_16214854 | 5.68 |
ENSDART00000109672
ENSDART00000190093 |
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr23_+_18722715 | 5.67 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr1_+_45056371 | 5.57 |
ENSDART00000073689
ENSDART00000167309 |
btr01
|
bloodthirsty-related gene family, member 1 |
| chr17_-_2595736 | 5.56 |
ENSDART00000128797
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr7_-_41851605 | 5.52 |
ENSDART00000142981
|
mylk3
|
myosin light chain kinase 3 |
| chr21_+_11749097 | 5.51 |
ENSDART00000102408
ENSDART00000102404 |
ell2
|
elongation factor, RNA polymerase II, 2 |
| chr13_-_36582341 | 5.49 |
ENSDART00000137335
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr16_+_29509133 | 5.46 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr7_+_27317174 | 5.42 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr2_-_3437862 | 5.41 |
ENSDART00000053012
|
cyp8b1
|
cytochrome P450, family 8, subfamily B, polypeptide 1 |
| chr21_-_20840714 | 5.40 |
ENSDART00000144861
ENSDART00000139430 |
c6
|
complement component 6 |
| chr9_-_12575776 | 5.35 |
ENSDART00000128931
ENSDART00000182695 |
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr12_-_4540564 | 5.26 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr11_-_25213651 | 5.21 |
ENSDART00000097316
ENSDART00000152186 |
myh7ba
|
myosin, heavy chain 7B, cardiac muscle, beta a |
| chr10_-_1961930 | 5.14 |
ENSDART00000122446
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr3_+_23743139 | 5.13 |
ENSDART00000187409
|
hoxb3a
|
homeobox B3a |
| chr7_+_48761875 | 5.13 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr11_-_40457325 | 5.04 |
ENSDART00000128442
|
tnfrsf1b
|
tumor necrosis factor receptor superfamily, member 1B |
| chr23_-_24263474 | 5.03 |
ENSDART00000160312
|
hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr9_+_48007081 | 5.02 |
ENSDART00000060593
ENSDART00000099835 |
zgc:92380
|
zgc:92380 |
| chr7_+_1473929 | 5.00 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr8_-_39739627 | 4.99 |
ENSDART00000135422
ENSDART00000067844 |
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr10_-_8053385 | 4.96 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr23_-_21215520 | 4.95 |
ENSDART00000143206
|
megf6a
|
multiple EGF-like-domains 6a |
| chr3_+_21189766 | 4.92 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr22_-_5323482 | 4.88 |
ENSDART00000145785
|
s1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr3_+_2669813 | 4.86 |
ENSDART00000014205
|
CR388047.1
|
|
| chr10_-_8046764 | 4.82 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr11_-_37997419 | 4.80 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr22_-_20695237 | 4.79 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr23_-_21215311 | 4.76 |
ENSDART00000112424
|
megf6a
|
multiple EGF-like-domains 6a |
| chr13_+_30228077 | 4.72 |
ENSDART00000100813
ENSDART00000147729 ENSDART00000133404 |
rps24
|
ribosomal protein S24 |
| chr10_+_10788811 | 4.70 |
ENSDART00000101077
ENSDART00000139143 |
ptgdsa
|
prostaglandin D2 synthase a |
| chr22_+_15624371 | 4.67 |
ENSDART00000124868
|
lpl
|
lipoprotein lipase |
| chr1_+_57145072 | 4.67 |
ENSDART00000152776
|
si:ch73-94k4.4
|
si:ch73-94k4.4 |
| chr7_-_24047316 | 4.66 |
ENSDART00000184799
|
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr4_+_7841627 | 4.65 |
ENSDART00000037997
|
ucmaa
|
upper zone of growth plate and cartilage matrix associated a |
| chr3_+_23742868 | 4.57 |
ENSDART00000153512
|
hoxb3a
|
homeobox B3a |
| chr17_+_50701748 | 4.52 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr6_+_27338512 | 4.51 |
ENSDART00000155004
|
klhl30
|
kelch-like family member 30 |
| chr1_-_11642432 | 4.50 |
ENSDART00000139755
ENSDART00000115316 |
si:dkey-26i13.4
|
si:dkey-26i13.4 |
| chr7_+_48761646 | 4.48 |
ENSDART00000017467
|
acana
|
aggrecan a |
| chr8_-_38159805 | 4.46 |
ENSDART00000112331
ENSDART00000180006 |
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr20_+_29634653 | 4.45 |
ENSDART00000101556
|
asap2b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2b |
| chr16_-_16761164 | 4.41 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr7_-_24046999 | 4.40 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr7_-_20453661 | 4.40 |
ENSDART00000174001
|
ntn5
|
netrin 5 |
| chr2_+_29884363 | 4.39 |
ENSDART00000139247
|
si:ch211-207d6.2
|
si:ch211-207d6.2 |
| chr5_+_28830643 | 4.37 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr19_+_19761966 | 4.37 |
ENSDART00000163697
|
hoxa3a
|
homeobox A3a |
| chr11_-_141592 | 4.35 |
ENSDART00000092787
|
cdk4
|
cyclin-dependent kinase 4 |
| chr7_+_13988075 | 4.34 |
ENSDART00000186812
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr23_+_19790962 | 4.33 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr8_-_38201415 | 4.32 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr14_+_41345175 | 4.24 |
ENSDART00000086104
|
nox1
|
NADPH oxidase 1 |
| chr19_-_15229421 | 4.23 |
ENSDART00000055619
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr7_+_30626378 | 4.20 |
ENSDART00000173533
ENSDART00000052541 |
ccnb2
|
cyclin B2 |
| chr12_-_4346085 | 4.17 |
ENSDART00000112433
|
ca15c
|
carbonic anhydrase XV c |
| chr9_-_9415000 | 4.14 |
ENSDART00000146210
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr1_+_57187794 | 4.11 |
ENSDART00000152485
|
si:dkey-27j5.9
|
si:dkey-27j5.9 |
| chr17_-_2590222 | 4.10 |
ENSDART00000185711
|
CR759892.1
|
|
| chr1_+_1805294 | 4.08 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 3 |
| chr20_+_38458084 | 4.06 |
ENSDART00000020153
ENSDART00000135912 |
coq8a
|
coenzyme Q8A |
| chr5_-_38197080 | 4.05 |
ENSDART00000140708
|
si:ch211-284e13.9
|
si:ch211-284e13.9 |
| chr17_-_30652738 | 4.05 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr17_+_15788100 | 4.04 |
ENSDART00000027667
|
rragd
|
ras-related GTP binding D |
| chr4_-_1147655 | 3.95 |
ENSDART00000171561
|
si:ch211-117i20.2
|
si:ch211-117i20.2 |
| chr15_-_452347 | 3.95 |
ENSDART00000115233
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr14_+_23520986 | 3.92 |
ENSDART00000170473
ENSDART00000175970 |
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr7_-_37919475 | 3.90 |
ENSDART00000052368
|
heatr3
|
HEAT repeat containing 3 |
| chr23_+_5631381 | 3.89 |
ENSDART00000149143
|
pkp1a
|
plakophilin 1a |
| chr19_-_3777217 | 3.86 |
ENSDART00000160510
|
si:dkey-206d17.15
|
si:dkey-206d17.15 |
| chr10_+_41199660 | 3.85 |
ENSDART00000125314
|
adrb3b
|
adrenoceptor beta 3b |
| chr7_-_16598212 | 3.85 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr6_+_52927651 | 3.84 |
ENSDART00000141094
|
si:dkeyp-3f10.11
|
si:dkeyp-3f10.11 |
| chr23_-_3758637 | 3.84 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr12_-_36268723 | 3.83 |
ENSDART00000113740
|
kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr18_+_808911 | 3.82 |
ENSDART00000172518
|
cox5ab
|
cytochrome c oxidase subunit Vab |
| chr16_-_22192006 | 3.80 |
ENSDART00000163338
|
il6r
|
interleukin 6 receptor |
| chr8_-_32803227 | 3.77 |
ENSDART00000110079
|
zgc:194839
|
zgc:194839 |
| chr8_-_36399884 | 3.76 |
ENSDART00000108538
|
si:zfos-2070c2.3
|
si:zfos-2070c2.3 |
| chr1_-_58664854 | 3.71 |
ENSDART00000109528
|
adgre5b.3
|
adhesion G protein-coupled receptor E5b, duplicate 3 |
| chr8_+_6954984 | 3.69 |
ENSDART00000145610
|
si:ch211-255g12.6
|
si:ch211-255g12.6 |
| chr20_-_29418620 | 3.68 |
ENSDART00000172634
|
ryr3
|
ryanodine receptor 3 |
| chr16_+_54248706 | 3.62 |
ENSDART00000156915
|
drd2l
|
dopamine receptor D2 like |
| chr16_-_45152169 | 3.61 |
ENSDART00000171452
|
si:ch73-343g19.4
|
si:ch73-343g19.4 |
| chr21_-_13690712 | 3.61 |
ENSDART00000065817
|
pou5f3
|
POU domain, class 5, transcription factor 3 |
| chr5_-_44286987 | 3.59 |
ENSDART00000184112
|
si:ch73-337l15.2
|
si:ch73-337l15.2 |
| chr17_+_25833947 | 3.58 |
ENSDART00000044328
ENSDART00000154604 |
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr2_-_20599315 | 3.53 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr16_+_3982590 | 3.51 |
ENSDART00000149295
|
zc3h12a
|
zinc finger CCCH-type containing 12A |
| chr4_+_22480169 | 3.50 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr14_+_22022441 | 3.46 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr3_+_5087787 | 3.46 |
ENSDART00000162787
|
si:ch73-338o16.4
|
si:ch73-338o16.4 |
| chr8_+_32402441 | 3.45 |
ENSDART00000191451
|
epgn
|
epithelial mitogen homolog (mouse) |
| chr11_+_19159083 | 3.43 |
ENSDART00000138964
|
adamts9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr13_+_40501455 | 3.42 |
ENSDART00000114985
|
hpse2
|
heparanase 2 |
| chr11_+_24703108 | 3.42 |
ENSDART00000159173
|
gpr25
|
G protein-coupled receptor 25 |
| chr23_+_9560797 | 3.41 |
ENSDART00000180014
|
adrm1
|
adhesion regulating molecule 1 |
| chr11_+_36675599 | 3.40 |
ENSDART00000170464
|
si:ch211-11c3.11
|
si:ch211-11c3.11 |
| chr15_-_32383340 | 3.39 |
ENSDART00000185632
|
c4
|
complement component 4 |
| chr5_+_9408901 | 3.39 |
ENSDART00000193364
|
FP236810.1
|
|
| chr13_-_22961605 | 3.38 |
ENSDART00000143112
ENSDART00000057641 |
tspan15
|
tetraspanin 15 |
| chr4_-_64703 | 3.38 |
ENSDART00000167851
|
CU856344.1
|
|
| chr12_-_11258404 | 3.35 |
ENSDART00000149229
|
si:ch73-30l9.1
|
si:ch73-30l9.1 |
| chr18_+_39649825 | 3.34 |
ENSDART00000087960
|
gldn
|
gliomedin |
| chr17_-_114121 | 3.33 |
ENSDART00000172408
ENSDART00000157784 |
arhgap11a
|
Rho GTPase activating protein 11A |
| chr12_-_32421046 | 3.30 |
ENSDART00000075567
|
enpp7.1
|
ectonucleotide pyrophosphatase/phosphodiesterase 7, tandem duplicate 1 |
| chr16_-_35952789 | 3.30 |
ENSDART00000180118
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr9_-_51563575 | 3.27 |
ENSDART00000167034
ENSDART00000148918 |
tank
|
TRAF family member-associated NFKB activator |
| chr4_+_37936839 | 3.26 |
ENSDART00000164505
|
si:dkeyp-82b4.2
|
si:dkeyp-82b4.2 |
| chr5_+_9377005 | 3.26 |
ENSDART00000124924
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr18_+_39649660 | 3.26 |
ENSDART00000149859
|
gldn
|
gliomedin |
| chr5_-_34921046 | 3.25 |
ENSDART00000180245
|
arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr5_-_71838520 | 3.24 |
ENSDART00000174396
|
CU927890.1
|
|
| chr1_+_524388 | 3.23 |
ENSDART00000020327
|
mrpl16
|
mitochondrial ribosomal protein L16 |
| chr13_+_21870269 | 3.22 |
ENSDART00000144612
|
zswim8
|
zinc finger, SWIM-type containing 8 |
| chr13_-_290377 | 3.22 |
ENSDART00000134963
|
chs1
|
chitin synthase 1 |
| chr7_+_49681040 | 3.20 |
ENSDART00000176372
ENSDART00000192172 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr3_+_21200763 | 3.18 |
ENSDART00000067841
|
zgc:112038
|
zgc:112038 |
| chr20_-_25533739 | 3.17 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr4_+_72797711 | 3.17 |
ENSDART00000190934
ENSDART00000163236 |
MYRFL
|
myelin regulatory factor-like |
| chr21_-_21828261 | 3.16 |
ENSDART00000151181
|
defbl2
|
defensin, beta-like 2 |
| chr1_+_524717 | 3.12 |
ENSDART00000102421
ENSDART00000184473 |
mrpl16
|
mitochondrial ribosomal protein L16 |
| chr19_+_19756425 | 3.11 |
ENSDART00000167606
|
hoxa3a
|
homeobox A3a |
| chr14_+_16151636 | 3.10 |
ENSDART00000159352
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr10_+_31953502 | 3.10 |
ENSDART00000185634
|
lhfpl6
|
LHFPL tetraspan subfamily member 6 |
| chr4_+_16715267 | 3.08 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr15_+_37986069 | 3.07 |
ENSDART00000156984
|
si:dkey-238d18.8
|
si:dkey-238d18.8 |
| chr18_+_3004572 | 3.06 |
ENSDART00000190328
|
CABZ01002690.1
|
|
| chr8_-_38355555 | 3.05 |
ENSDART00000134283
ENSDART00000132077 ENSDART00000146378 |
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr1_+_12177195 | 3.04 |
ENSDART00000146842
ENSDART00000142081 |
stra6l
|
STRA6-like |
| chr23_-_45897900 | 3.03 |
ENSDART00000149263
|
ftr92
|
finTRIM family, member 92 |
| chr23_+_39963599 | 3.03 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr3_+_57820913 | 2.99 |
ENSDART00000168101
|
CU571328.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa9a+hoxa9b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 18.7 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 2.7 | 8.0 | GO:0090008 | hypoblast development(GO:0090008) |
| 2.4 | 7.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 2.3 | 16.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 2.3 | 6.8 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 2.1 | 6.3 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 1.9 | 9.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 1.8 | 7.3 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 1.8 | 10.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 1.7 | 7.0 | GO:0019626 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 1.6 | 8.2 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 1.6 | 9.6 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 1.5 | 4.5 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 1.5 | 7.3 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 1.4 | 5.6 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 1.2 | 7.2 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 1.1 | 5.5 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 1.0 | 7.0 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 1.0 | 2.9 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 1.0 | 3.8 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.9 | 5.7 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.9 | 5.5 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.9 | 2.7 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.9 | 3.5 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.9 | 4.3 | GO:0071072 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.9 | 8.6 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.8 | 2.5 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.8 | 5.0 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.8 | 2.4 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.8 | 3.8 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.8 | 2.3 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.8 | 7.6 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.7 | 12.6 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.7 | 14.8 | GO:0014823 | response to activity(GO:0014823) |
| 0.7 | 2.7 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.7 | 2.0 | GO:0015882 | L-ascorbic acid transport(GO:0015882) |
| 0.7 | 5.9 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.6 | 3.2 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.6 | 11.3 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.6 | 3.0 | GO:0035461 | vitamin transmembrane transport(GO:0035461) vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.6 | 3.0 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.6 | 4.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.6 | 1.8 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.6 | 3.6 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.6 | 2.3 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.6 | 2.8 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.6 | 7.8 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.6 | 2.2 | GO:0032965 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.5 | 3.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.5 | 5.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.5 | 4.2 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.5 | 2.1 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.5 | 2.0 | GO:0015677 | copper ion import(GO:0015677) |
| 0.5 | 3.5 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.5 | 2.9 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.5 | 2.4 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.5 | 6.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.5 | 3.6 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.5 | 22.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.4 | 1.8 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.4 | 4.8 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.4 | 4.3 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.4 | 2.5 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.4 | 1.2 | GO:0071435 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 0.4 | 2.4 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.4 | 4.6 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.4 | 5.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.4 | 3.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.4 | 2.3 | GO:1901655 | cellular response to ketone(GO:1901655) |
| 0.4 | 14.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.3 | 2.4 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.3 | 1.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.3 | 5.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.3 | 3.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.3 | 5.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.3 | 5.0 | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.3 | 15.1 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.3 | 1.8 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.3 | 1.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 1.8 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.3 | 3.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 | 2.5 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.3 | 7.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.3 | 1.0 | GO:1903798 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.2 | 2.7 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.2 | 1.7 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 3.1 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.2 | 1.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 2.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.2 | 1.0 | GO:2000562 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 | 1.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.2 | 3.0 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 1.5 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.2 | 4.1 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.2 | 2.8 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.2 | 6.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.2 | 9.8 | GO:0007602 | phototransduction(GO:0007602) |
| 0.2 | 4.5 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.2 | 1.1 | GO:0006415 | translational termination(GO:0006415) |
| 0.2 | 9.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.2 | 1.6 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.2 | 4.0 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.2 | 1.5 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 1.2 | GO:0035912 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.1 | 2.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 1.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 3.4 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 7.8 | GO:0006354 | DNA-templated transcription, elongation(GO:0006354) |
| 0.1 | 39.2 | GO:0060537 | muscle tissue development(GO:0060537) |
| 0.1 | 2.9 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 10.7 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 6.6 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 2.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 1.8 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 11.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.6 | GO:0010888 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.1 | 0.8 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.1 | 0.7 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 1.1 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.1 | 0.7 | GO:0044319 | wound healing, spreading of cells(GO:0044319) epiboly involved in wound healing(GO:0090505) skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 5.1 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.1 | 0.4 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 1.7 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 1.2 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 2.0 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.1 | 2.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 1.9 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 4.9 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 8.4 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 2.0 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 1.1 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 0.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 17.2 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 7.2 | GO:0051216 | cartilage development(GO:0051216) |
| 0.1 | 6.1 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.1 | 0.5 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 0.9 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 | 0.6 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 1.7 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 0.3 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 4.7 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.1 | 3.2 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 1.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 1.2 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.1 | 13.5 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 2.9 | GO:2000027 | regulation of organ morphogenesis(GO:2000027) |
| 0.1 | 0.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 1.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 1.7 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 2.0 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 1.2 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 17.3 | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway(GO:0007169) |
| 0.0 | 0.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 1.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 1.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 1.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 2.9 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 2.9 | GO:0001666 | response to hypoxia(GO:0001666) response to decreased oxygen levels(GO:0036293) |
| 0.0 | 0.7 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 1.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 12.5 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 2.1 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 2.0 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 1.1 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 1.1 | GO:0031076 | embryonic camera-type eye development(GO:0031076) |
| 0.0 | 2.4 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 1.7 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 2.7 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 3.4 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 0.4 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 1.4 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 2.2 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.0 | 1.0 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 1.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) regulation of B cell activation(GO:0050864) positive regulation of B cell activation(GO:0050871) |
| 0.0 | 0.1 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 1.4 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.0 | 1.1 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.1 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.3 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 8.8 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.0 | 3.0 | GO:0006644 | phospholipid metabolic process(GO:0006644) |
| 0.0 | 5.8 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.9 | GO:0008203 | cholesterol metabolic process(GO:0008203) secondary alcohol metabolic process(GO:1902652) |
| 0.0 | 1.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.6 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 1.3 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 1.0 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 1.0 | 7.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 1.0 | 4.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.8 | 4.0 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.7 | 5.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.6 | 1.8 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.6 | 7.6 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.6 | 2.3 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.5 | 4.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.5 | 5.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.5 | 3.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.5 | 22.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.4 | 15.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.4 | 3.2 | GO:0030428 | cell septum(GO:0030428) |
| 0.4 | 4.7 | GO:0042627 | chylomicron(GO:0042627) |
| 0.4 | 45.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.3 | 1.4 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.3 | 0.9 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.3 | 1.8 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.3 | 2.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.3 | 3.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 2.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 3.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 3.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 13.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 2.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 3.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.2 | 2.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 8.9 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.2 | 2.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 17.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 23.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.2 | 2.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 60.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 1.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 6.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.8 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 2.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 5.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.9 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 3.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 14.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 4.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 1.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 1.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 2.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 5.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 3.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 24.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 1.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 5.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.4 | GO:0098798 | mitochondrial protein complex(GO:0098798) |
| 0.0 | 1.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 4.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 4.3 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 2.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 3.6 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 2.5 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 6.5 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 3.5 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 1.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 2.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 4.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 3.6 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 2.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 14.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.1 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 7.0 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.7 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 2.1 | 8.6 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 1.8 | 5.4 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 1.7 | 18.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.6 | 4.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 1.5 | 15.5 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 1.3 | 6.5 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 1.1 | 5.5 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 1.0 | 5.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 1.0 | 3.8 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.9 | 5.7 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.8 | 7.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.8 | 2.4 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.7 | 3.7 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.7 | 15.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.7 | 3.4 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.7 | 2.7 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.7 | 4.6 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.6 | 8.0 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.6 | 4.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.6 | 4.9 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.6 | 3.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.6 | 2.9 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.6 | 2.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.6 | 6.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.5 | 3.8 | GO:0005549 | olfactory receptor activity(GO:0004984) odorant binding(GO:0005549) |
| 0.5 | 2.7 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.5 | 7.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.5 | 3.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.5 | 2.0 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.5 | 5.6 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.5 | 15.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.5 | 3.8 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.5 | 4.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.4 | 3.0 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.4 | 5.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.4 | 3.2 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.4 | 1.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.4 | 3.0 | GO:0015157 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.3 | 11.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 1.6 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.3 | 4.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 1.9 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.3 | 4.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.3 | 2.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 2.8 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.3 | 7.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.3 | 2.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 2.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 13.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.3 | 1.8 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.2 | 14.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 1.7 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 6.3 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.2 | 3.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 4.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 4.2 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.2 | 9.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 4.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.2 | 17.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 2.8 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 9.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.2 | 7.0 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.2 | 0.6 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.2 | 4.1 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 0.9 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.2 | 2.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 0.9 | GO:0016415 | octanoyltransferase activity(GO:0016415) |
| 0.2 | 1.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.2 | 3.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 24.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 4.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 3.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 10.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.4 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.1 | 1.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.4 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 3.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 2.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 3.5 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 6.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 1.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 2.9 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 8.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 2.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 1.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 2.9 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 8.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 16.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 17.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 4.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 2.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 2.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.4 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 1.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 2.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 19.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 8.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 4.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.7 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 2.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 1.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 2.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 3.5 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 6.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.8 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 14.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 0.2 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 2.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 8.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 5.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.4 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 3.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 15.8 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 6.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 1.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 1.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 12.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.4 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 2.3 | GO:0016798 | hydrolase activity, acting on glycosyl bonds(GO:0016798) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 18.6 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 3.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.8 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 8.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.4 | 14.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 4.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.3 | 7.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.3 | 8.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.3 | 4.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 11.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 5.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 8.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 30.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 7.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 2.8 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 1.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 4.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 3.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 4.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 2.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 7.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 6.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 14.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 1.1 | 5.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 1.0 | 5.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.8 | 8.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.7 | 3.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.7 | 9.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.6 | 7.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.5 | 11.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.5 | 4.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.4 | 5.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.4 | 5.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.4 | 2.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 3.8 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.4 | 4.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.3 | 2.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 7.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.3 | 5.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 4.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 3.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 2.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 4.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 2.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.2 | 3.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 3.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 18.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 4.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 0.7 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 7.3 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 3.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 0.5 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 3.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 6.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 2.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.2 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 3.9 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 1.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 2.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |