Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
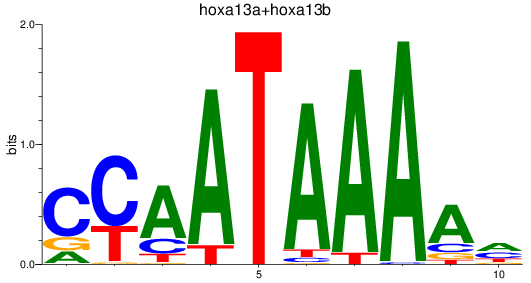
Results for hoxa13a+hoxa13b
Z-value: 1.51
Transcription factors associated with hoxa13a+hoxa13b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa13b
|
ENSDARG00000036254 | homeobox A13b |
|
hoxa13a
|
ENSDARG00000100312 | homeobox A13a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa13a | dr11_v1_chr19_+_19729506_19729506 | 0.07 | 5.3e-01 | Click! |
| hoxa13b | dr11_v1_chr16_+_20895904_20895904 | 0.01 | 9.6e-01 | Click! |
Activity profile of hoxa13a+hoxa13b motif
Sorted Z-values of hoxa13a+hoxa13b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_30611814 | 11.55 |
ENSDART00000089803
|
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr2_-_24289641 | 11.04 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr17_-_15382704 | 9.02 |
ENSDART00000005313
|
zgc:85722
|
zgc:85722 |
| chr2_+_16781015 | 7.95 |
ENSDART00000155147
ENSDART00000003845 |
tfa
|
transferrin-a |
| chr14_+_36738069 | 7.83 |
ENSDART00000105590
|
tdo2a
|
tryptophan 2,3-dioxygenase a |
| chr2_+_16780643 | 6.91 |
ENSDART00000125647
ENSDART00000108611 ENSDART00000181245 ENSDART00000163194 |
tfa
|
transferrin-a |
| chr16_-_21785261 | 6.87 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr23_+_7379728 | 6.57 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr21_-_2707768 | 5.99 |
ENSDART00000165384
|
CABZ01101739.1
|
|
| chr21_-_25756119 | 5.91 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr7_-_16562200 | 5.86 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr13_+_23132666 | 5.65 |
ENSDART00000164639
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr1_+_55140970 | 5.63 |
ENSDART00000039807
|
mb
|
myoglobin |
| chr22_-_24818066 | 4.95 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr5_-_42071505 | 4.69 |
ENSDART00000137224
ENSDART00000193721 |
cxcl11.7
cenpv
|
chemokine (C-X-C motif) ligand 11, duplicate 7 centromere protein V |
| chr4_+_7391110 | 4.50 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr11_+_24001993 | 4.47 |
ENSDART00000168215
|
chia.2
|
chitinase, acidic.2 |
| chr14_-_21123551 | 4.34 |
ENSDART00000171679
ENSDART00000165882 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr11_+_24313931 | 4.33 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr4_+_7391400 | 4.31 |
ENSDART00000169111
ENSDART00000186395 |
tnni4a
|
troponin I4a |
| chr7_-_26076970 | 4.11 |
ENSDART00000101120
|
zgc:92664
|
zgc:92664 |
| chr5_-_42883761 | 3.95 |
ENSDART00000167374
|
BX323596.2
|
|
| chr22_+_15624371 | 3.93 |
ENSDART00000124868
|
lpl
|
lipoprotein lipase |
| chr13_+_24584401 | 3.93 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr10_+_17026870 | 3.93 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr23_+_22200467 | 3.74 |
ENSDART00000025414
|
slc2a1a
|
solute carrier family 2 (facilitated glucose transporter), member 1a |
| chr16_-_45917322 | 3.74 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr25_+_13620555 | 3.68 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr16_+_46459680 | 3.67 |
ENSDART00000101698
|
rpz3
|
rapunzel 3 |
| chr11_-_3343463 | 3.64 |
ENSDART00000066177
|
tuba2
|
tubulin, alpha 2 |
| chr11_-_24016761 | 3.63 |
ENSDART00000153601
ENSDART00000067817 ENSDART00000170531 |
chia.3
|
chitinase, acidic.3 |
| chr17_-_20897250 | 3.62 |
ENSDART00000088106
|
ank3b
|
ankyrin 3b |
| chr11_+_24002503 | 3.57 |
ENSDART00000164702
|
chia.2
|
chitinase, acidic.2 |
| chr2_+_21855036 | 3.50 |
ENSDART00000140012
|
ca8
|
carbonic anhydrase VIII |
| chr13_-_37619159 | 3.35 |
ENSDART00000186348
|
zgc:152791
|
zgc:152791 |
| chr13_+_844150 | 3.27 |
ENSDART00000058260
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr11_+_6819050 | 3.27 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr12_+_27704015 | 3.26 |
ENSDART00000153256
|
cacna1g
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr7_-_30367650 | 3.23 |
ENSDART00000075519
|
aldh1a2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr8_+_30709685 | 3.19 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr8_+_31821396 | 3.14 |
ENSDART00000077053
|
plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr11_+_24314148 | 3.09 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr6_-_53426773 | 3.04 |
ENSDART00000162791
|
mst1
|
macrophage stimulating 1 |
| chr14_+_31618982 | 3.01 |
ENSDART00000026195
|
slc9a6a
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6a |
| chr6_+_37301341 | 2.99 |
ENSDART00000104180
|
zranb2
|
zinc finger, RAN-binding domain containing 2 |
| chr3_-_24980067 | 2.99 |
ENSDART00000048871
|
desi1a
|
desumoylating isopeptidase 1a |
| chr2_-_32624577 | 2.97 |
ENSDART00000112797
|
si:dkeyp-73d8.6
|
si:dkeyp-73d8.6 |
| chr23_-_18030399 | 2.96 |
ENSDART00000136967
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr1_+_14073891 | 2.91 |
ENSDART00000021693
|
ank2a
|
ankyrin 2a, neuronal |
| chr8_+_30671060 | 2.91 |
ENSDART00000193749
|
adora2aa
|
adenosine A2a receptor a |
| chr21_-_20939488 | 2.91 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr15_+_25683069 | 2.87 |
ENSDART00000148190
|
hic1
|
hypermethylated in cancer 1 |
| chr11_-_44484952 | 2.83 |
ENSDART00000166674
ENSDART00000188016 |
mfn1b
|
mitofusin 1b |
| chr3_-_37759969 | 2.83 |
ENSDART00000151105
ENSDART00000151208 |
si:dkey-260c8.6
|
si:dkey-260c8.6 |
| chr15_-_21132480 | 2.81 |
ENSDART00000078734
ENSDART00000157481 |
a2ml
|
alpha-2-macroglobulin-like |
| chr22_+_1911269 | 2.80 |
ENSDART00000164158
ENSDART00000168205 |
znf1156
|
zinc finger protein 1156 |
| chr5_+_21211135 | 2.79 |
ENSDART00000088492
|
bmp10
|
bone morphogenetic protein 10 |
| chr13_-_24448278 | 2.79 |
ENSDART00000057584
|
slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr2_+_10147029 | 2.77 |
ENSDART00000139064
ENSDART00000053426 ENSDART00000153678 |
pfn2l
|
profilin 2 like |
| chr4_+_14900042 | 2.76 |
ENSDART00000018261
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr12_+_26471712 | 2.71 |
ENSDART00000162115
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr7_+_37372479 | 2.70 |
ENSDART00000173652
|
sall1a
|
spalt-like transcription factor 1a |
| chr2_+_23006792 | 2.66 |
ENSDART00000027782
|
mknk2a
|
MAP kinase interacting serine/threonine kinase 2a |
| chr12_-_14211293 | 2.66 |
ENSDART00000158399
|
avl9
|
AVL9 homolog (S. cerevisiase) |
| chr24_+_25692802 | 2.66 |
ENSDART00000190493
|
camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr2_+_35595454 | 2.65 |
ENSDART00000098734
|
cacybp
|
calcyclin binding protein |
| chr5_-_14390445 | 2.65 |
ENSDART00000026120
|
ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr6_-_60147517 | 2.64 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr2_-_54639964 | 2.63 |
ENSDART00000100103
|
acss2l
|
acyl-CoA synthetase short chain family member 2 like |
| chr4_-_16836006 | 2.58 |
ENSDART00000010777
|
ldhba
|
lactate dehydrogenase Ba |
| chr12_-_31457301 | 2.57 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr9_-_38036984 | 2.55 |
ENSDART00000134574
|
hacd2
|
3-hydroxyacyl-CoA dehydratase 2 |
| chr13_-_8692432 | 2.53 |
ENSDART00000058106
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr15_+_45586471 | 2.49 |
ENSDART00000165699
|
cldn15lb
|
claudin 15-like b |
| chr22_-_38819603 | 2.48 |
ENSDART00000104437
|
si:ch211-262h13.5
|
si:ch211-262h13.5 |
| chr20_+_32501748 | 2.48 |
ENSDART00000152944
ENSDART00000021035 |
sec63
|
SEC63 homolog, protein translocation regulator |
| chr3_+_46764278 | 2.44 |
ENSDART00000136051
ENSDART00000164930 |
prkcsh
|
protein kinase C substrate 80K-H |
| chr7_+_38507006 | 2.41 |
ENSDART00000173830
|
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr25_+_34641536 | 2.41 |
ENSDART00000167033
|
CABZ01079011.1
|
|
| chr15_-_12500938 | 2.40 |
ENSDART00000159627
|
scn4ba
|
sodium channel, voltage-gated, type IV, beta a |
| chr4_+_5156117 | 2.38 |
ENSDART00000067392
|
tigarb
|
tp53-induced glycolysis and apoptosis regulator b |
| chr10_-_25823258 | 2.36 |
ENSDART00000064327
|
ftr54
|
finTRIM family, member 54 |
| chr4_+_45148652 | 2.35 |
ENSDART00000150798
|
si:dkey-51d8.9
|
si:dkey-51d8.9 |
| chr17_-_52587598 | 2.34 |
ENSDART00000061497
|
si:ch211-173a9.6
|
si:ch211-173a9.6 |
| chr1_-_9249943 | 2.34 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr3_+_1005059 | 2.33 |
ENSDART00000132460
|
zgc:172051
|
zgc:172051 |
| chr16_+_42464613 | 2.32 |
ENSDART00000162454
|
si:ch211-215k15.4
|
si:ch211-215k15.4 |
| chr13_-_11986754 | 2.32 |
ENSDART00000164214
|
npm3
|
nucleophosmin/nucleoplasmin, 3 |
| chr13_+_25380432 | 2.32 |
ENSDART00000038524
|
gsto1
|
glutathione S-transferase omega 1 |
| chr17_-_12389259 | 2.31 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr23_+_18722715 | 2.29 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr17_+_43032529 | 2.27 |
ENSDART00000055611
ENSDART00000154863 |
isca2
|
iron-sulfur cluster assembly 2 |
| chr17_-_7371564 | 2.26 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr16_-_563235 | 2.24 |
ENSDART00000016303
|
irx2a
|
iroquois homeobox 2a |
| chr22_-_16377960 | 2.23 |
ENSDART00000168170
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr2_+_21855291 | 2.22 |
ENSDART00000186204
|
ca8
|
carbonic anhydrase VIII |
| chr21_-_37733287 | 2.21 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr21_+_22878991 | 2.20 |
ENSDART00000186399
|
pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr22_-_12160283 | 2.20 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr23_+_18722915 | 2.20 |
ENSDART00000025057
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr5_-_41307550 | 2.18 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr7_+_48555400 | 2.17 |
ENSDART00000174474
|
kcnq1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr16_+_23921610 | 2.17 |
ENSDART00000143855
|
apoa4b.3
|
apolipoprotein A-IV b, tandem duplicate 3 |
| chr17_-_43031763 | 2.16 |
ENSDART00000132754
ENSDART00000050399 |
npc2
|
Niemann-Pick disease, type C2 |
| chr9_+_32301017 | 2.16 |
ENSDART00000127916
ENSDART00000183298 ENSDART00000143103 |
hspe1
|
heat shock 10 protein 1 |
| chr17_+_15788100 | 2.16 |
ENSDART00000027667
|
rragd
|
ras-related GTP binding D |
| chr17_+_31820401 | 2.15 |
ENSDART00000192607
ENSDART00000157490 |
eef1akmt2
|
EEF1A lysine methyltransferase 2 |
| chr14_+_33722950 | 2.14 |
ENSDART00000075312
|
apln
|
apelin |
| chr12_+_41697664 | 2.13 |
ENSDART00000162302
|
bnip3
|
BCL2 interacting protein 3 |
| chr7_-_26457208 | 2.13 |
ENSDART00000173519
|
zgc:172079
|
zgc:172079 |
| chr24_-_23671383 | 2.11 |
ENSDART00000144193
|
hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr1_+_53377432 | 2.10 |
ENSDART00000177581
|
ucp1
|
uncoupling protein 1 |
| chr25_-_31739309 | 2.10 |
ENSDART00000098896
|
acot19
|
acyl-CoA thioesterase 19 |
| chr5_+_41143563 | 2.10 |
ENSDART00000011229
|
sub1b
|
SUB1 homolog, transcriptional regulator b |
| chr10_-_26744131 | 2.09 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr13_+_28675686 | 2.09 |
ENSDART00000027213
|
inaa
|
internexin neuronal intermediate filament protein, alpha a |
| chr15_+_14856307 | 2.09 |
ENSDART00000167213
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr8_+_14886452 | 2.08 |
ENSDART00000146589
|
soat1
|
sterol O-acyltransferase 1 |
| chr9_+_32301456 | 2.07 |
ENSDART00000078608
ENSDART00000185153 ENSDART00000144947 |
hspe1
|
heat shock 10 protein 1 |
| chr2_-_51757328 | 2.07 |
ENSDART00000189286
|
si:ch211-9d9.1
|
si:ch211-9d9.1 |
| chr8_+_40477264 | 2.07 |
ENSDART00000085559
|
gck
|
glucokinase (hexokinase 4) |
| chr12_+_9703172 | 2.05 |
ENSDART00000091489
|
ppp1r9bb
|
protein phosphatase 1, regulatory subunit 9Bb |
| chr8_+_14381272 | 2.03 |
ENSDART00000057642
|
acbd6
|
acyl-CoA binding domain containing 6 |
| chr6_-_40697585 | 2.02 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr6_+_46431848 | 2.02 |
ENSDART00000181056
ENSDART00000144569 ENSDART00000064865 ENSDART00000133992 |
stau1
|
staufen double-stranded RNA binding protein 1 |
| chr14_-_32959851 | 2.00 |
ENSDART00000075157
|
chic1
|
cysteine-rich hydrophobic domain 1 |
| chr25_+_336503 | 1.98 |
ENSDART00000160395
|
CU929262.1
|
|
| chr17_-_14451718 | 1.96 |
ENSDART00000039438
|
jkamp
|
jnk1/mapk8 associated membrane protein |
| chr24_-_23671709 | 1.95 |
ENSDART00000164750
|
hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr8_+_28467893 | 1.94 |
ENSDART00000189724
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr7_-_20865005 | 1.94 |
ENSDART00000190752
|
fis1
|
fission, mitochondrial 1 |
| chr7_+_41314862 | 1.94 |
ENSDART00000185198
|
zgc:165532
|
zgc:165532 |
| chr13_+_28690355 | 1.94 |
ENSDART00000137475
ENSDART00000128246 |
polr1c
|
polymerase (RNA) I polypeptide C |
| chr3_+_16612574 | 1.94 |
ENSDART00000104481
|
slc17a7a
|
solute carrier family 17 (vesicular glutamate transporter), member 7a |
| chr24_-_6158933 | 1.93 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr9_+_38074082 | 1.92 |
ENSDART00000017833
|
cacnb4a
|
calcium channel, voltage-dependent, beta 4a subunit |
| chr25_+_28823952 | 1.91 |
ENSDART00000067072
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr21_-_25748913 | 1.91 |
ENSDART00000134217
|
CR388166.2
|
|
| chr8_-_16738204 | 1.90 |
ENSDART00000100706
|
ndufaf2
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 2 |
| chr9_-_32300783 | 1.90 |
ENSDART00000078596
|
hspd1
|
heat shock 60 protein 1 |
| chr10_-_9269733 | 1.89 |
ENSDART00000064974
|
anxa3b
|
annexin A3b |
| chr22_+_24715282 | 1.87 |
ENSDART00000088027
ENSDART00000189054 ENSDART00000140430 |
ssx2ipb
|
synovial sarcoma, X breakpoint 2 interacting protein b |
| chr14_-_12837432 | 1.86 |
ENSDART00000178444
|
gria3b
|
glutamate receptor, ionotropic, AMPA 3b |
| chr12_+_19199735 | 1.86 |
ENSDART00000066393
|
pdap1a
|
pdgfa associated protein 1a |
| chr5_-_37881345 | 1.85 |
ENSDART00000084819
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr25_-_19374710 | 1.83 |
ENSDART00000184483
ENSDART00000188706 |
map1ab
|
microtubule-associated protein 1Ab |
| chr13_+_4405282 | 1.82 |
ENSDART00000148280
|
prr18
|
proline rich 18 |
| chr22_+_21972824 | 1.82 |
ENSDART00000183708
ENSDART00000133739 |
gna15.3
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 3 |
| chr18_+_7283283 | 1.82 |
ENSDART00000141493
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr4_+_5506952 | 1.82 |
ENSDART00000032857
ENSDART00000160222 |
mapk11
|
mitogen-activated protein kinase 11 |
| chr25_-_12906872 | 1.81 |
ENSDART00000165156
ENSDART00000167449 |
sept15
|
septin 15 |
| chr8_+_38564942 | 1.81 |
ENSDART00000085371
|
rfk
|
riboflavin kinase |
| chr14_+_35748385 | 1.80 |
ENSDART00000064617
ENSDART00000074671 ENSDART00000172803 |
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr7_-_44604821 | 1.80 |
ENSDART00000148967
|
tk2
|
thymidine kinase 2, mitochondrial |
| chr23_-_21446985 | 1.80 |
ENSDART00000044080
|
her12
|
hairy-related 12 |
| chr10_+_42678520 | 1.79 |
ENSDART00000182496
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr16_-_560574 | 1.79 |
ENSDART00000148452
|
irx2a
|
iroquois homeobox 2a |
| chr15_-_5901514 | 1.79 |
ENSDART00000155252
|
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr25_-_24046870 | 1.79 |
ENSDART00000047569
|
igf2b
|
insulin-like growth factor 2b |
| chr7_-_58098814 | 1.79 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr23_+_17417539 | 1.78 |
ENSDART00000182605
|
BX649300.2
|
|
| chr23_-_17484555 | 1.78 |
ENSDART00000187181
|
dnajc5ab
|
DnaJ (Hsp40) homolog, subfamily C, member 5ab |
| chr18_+_35861930 | 1.78 |
ENSDART00000185223
|
ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr14_+_35748206 | 1.78 |
ENSDART00000177391
|
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr19_-_7406933 | 1.77 |
ENSDART00000151137
|
oxr1b
|
oxidation resistance 1b |
| chr3_-_60316118 | 1.77 |
ENSDART00000171458
|
si:ch211-214b16.2
|
si:ch211-214b16.2 |
| chr2_+_31838442 | 1.76 |
ENSDART00000066789
|
stard3nl
|
STARD3 N-terminal like |
| chr18_+_16330720 | 1.75 |
ENSDART00000080638
|
nts
|
neurotensin |
| chr9_+_41080029 | 1.75 |
ENSDART00000141179
ENSDART00000019289 |
zgc:136439
|
zgc:136439 |
| chr10_+_22003750 | 1.73 |
ENSDART00000109420
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr6_+_43450221 | 1.72 |
ENSDART00000075521
|
zgc:113054
|
zgc:113054 |
| chr21_-_30111134 | 1.71 |
ENSDART00000014223
|
slc23a1
|
solute carrier family 23 (ascorbic acid transporter), member 1 |
| chr7_-_69025306 | 1.70 |
ENSDART00000180796
|
CABZ01057488.2
|
|
| chr7_+_71503356 | 1.70 |
ENSDART00000100396
|
ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr12_+_28367557 | 1.70 |
ENSDART00000066294
|
cdk5r1b
|
cyclin-dependent kinase 5, regulatory subunit 1b (p35) |
| chr17_-_31360951 | 1.70 |
ENSDART00000062894
ENSDART00000134686 |
ivd
|
isovaleryl-CoA dehydrogenase |
| chr7_-_39751540 | 1.69 |
ENSDART00000016803
|
grpel1
|
GrpE-like 1, mitochondrial |
| chr22_+_11857356 | 1.69 |
ENSDART00000179540
|
mras
|
muscle RAS oncogene homolog |
| chr19_-_41199944 | 1.68 |
ENSDART00000145135
|
pdk4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr10_-_19523752 | 1.68 |
ENSDART00000160057
|
si:ch211-127i16.2
|
si:ch211-127i16.2 |
| chr20_-_40487208 | 1.67 |
ENSDART00000075070
ENSDART00000142029 |
hsf2
|
heat shock transcription factor 2 |
| chr19_+_25526354 | 1.67 |
ENSDART00000052396
|
sdhaf3
|
succinate dehydrogenase complex assembly factor 3 |
| chr24_-_23675446 | 1.67 |
ENSDART00000066644
|
hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr5_-_51903243 | 1.67 |
ENSDART00000125535
|
mtx3
|
metaxin 3 |
| chr13_-_36844945 | 1.66 |
ENSDART00000129562
ENSDART00000150899 |
nin
|
ninein (GSK3B interacting protein) |
| chr10_+_8527196 | 1.66 |
ENSDART00000141147
|
si:ch211-193e13.5
|
si:ch211-193e13.5 |
| chr16_+_7626535 | 1.66 |
ENSDART00000182670
ENSDART00000065514 ENSDART00000150212 |
stx12l
|
syntaxin 12, like |
| chr21_+_40239192 | 1.65 |
ENSDART00000135857
|
or115-9
|
odorant receptor, family F, subfamily 115, member 9 |
| chr20_+_30490682 | 1.64 |
ENSDART00000184871
|
myt1la
|
myelin transcription factor 1-like, a |
| chr5_+_31946973 | 1.64 |
ENSDART00000189876
ENSDART00000163366 |
ungb
|
uracil DNA glycosylase b |
| chr9_-_32753535 | 1.64 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr10_+_22891126 | 1.63 |
ENSDART00000057291
|
arrb2a
|
arrestin, beta 2a |
| chr12_+_26706745 | 1.63 |
ENSDART00000141401
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr13_+_17694845 | 1.63 |
ENSDART00000079778
|
ifit8
|
interferon-induced protein with tetratricopeptide repeats 8 |
| chr8_+_40476811 | 1.62 |
ENSDART00000129772
|
gck
|
glucokinase (hexokinase 4) |
| chr16_+_32559821 | 1.62 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr8_+_19489854 | 1.61 |
ENSDART00000184671
ENSDART00000011258 |
npl
|
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase) |
| chr22_+_10713713 | 1.61 |
ENSDART00000122349
|
hiat1b
|
hippocampus abundant transcript 1b |
| chr2_+_30960351 | 1.61 |
ENSDART00000141575
|
lpin2
|
lipin 2 |
| chr11_-_29623380 | 1.61 |
ENSDART00000162587
ENSDART00000193935 ENSDART00000191646 |
chd5
|
chromodomain helicase DNA binding protein 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa13a+hoxa13b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.6 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 1.6 | 6.5 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 1.5 | 7.4 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 1.4 | 2.9 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 1.3 | 14.6 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 1.1 | 3.3 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 1.1 | 3.2 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 1.1 | 11.7 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 1.0 | 8.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 1.0 | 3.0 | GO:0015882 | L-ascorbic acid transport(GO:0015882) |
| 0.9 | 2.6 | GO:2000193 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.8 | 3.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.8 | 2.4 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 0.7 | 3.0 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.7 | 2.9 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.7 | 2.7 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.7 | 3.9 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.6 | 1.9 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.6 | 1.9 | GO:0006747 | FAD biosynthetic process(GO:0006747) flavin-containing compound metabolic process(GO:0042726) flavin-containing compound biosynthetic process(GO:0042727) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.6 | 3.2 | GO:0019482 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.6 | 1.2 | GO:0071635 | regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.6 | 4.6 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.6 | 2.8 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.5 | 2.9 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.5 | 1.4 | GO:0055026 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.5 | 3.7 | GO:0046323 | glucose import(GO:0046323) |
| 0.5 | 2.7 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.4 | 1.3 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.4 | 2.6 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.4 | 1.3 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.4 | 3.0 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.4 | 2.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.4 | 1.3 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.4 | 1.3 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.4 | 2.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.4 | 8.3 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.4 | 1.6 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.4 | 1.6 | GO:0097037 | heme export(GO:0097037) |
| 0.4 | 8.0 | GO:0014823 | response to activity(GO:0014823) |
| 0.4 | 1.6 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.4 | 0.8 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.4 | 1.9 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.4 | 3.7 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.4 | 1.1 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.4 | 2.2 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.4 | 1.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.4 | 1.8 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.4 | 2.1 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.4 | 2.5 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.4 | 2.5 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.3 | 1.4 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.3 | 1.4 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.3 | 1.7 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.3 | 1.6 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.3 | 2.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 1.9 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.3 | 1.6 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.3 | 1.6 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.3 | 4.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.3 | 0.9 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.3 | 0.9 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.3 | 0.9 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.3 | 1.7 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.3 | 3.0 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 1.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 2.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.3 | 1.6 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.3 | 1.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 1.2 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.2 | 1.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 0.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 1.7 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.2 | 3.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 2.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 2.1 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.2 | 1.2 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.2 | 3.9 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 2.1 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.2 | 0.7 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.2 | 0.9 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.2 | 0.9 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.2 | 0.9 | GO:0050960 | thermoception(GO:0050955) detection of temperature stimulus involved in thermoception(GO:0050960) detection of temperature stimulus involved in sensory perception(GO:0050961) |
| 0.2 | 0.6 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.2 | 0.8 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.2 | 3.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.2 | 0.8 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.2 | 1.4 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 2.8 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 1.5 | GO:0006111 | regulation of gluconeogenesis(GO:0006111) |
| 0.2 | 1.6 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.2 | 1.4 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.2 | 1.3 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.2 | 2.7 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.2 | 1.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 0.5 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 0.9 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.2 | 1.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 1.3 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.2 | 1.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 5.3 | GO:0051084 | 'de novo' protein folding(GO:0006458) 'de novo' posttranslational protein folding(GO:0051084) chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.2 | 1.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 1.0 | GO:0061193 | rhombomere 5 development(GO:0021571) rhombomere 6 development(GO:0021572) tongue development(GO:0043586) tongue morphogenesis(GO:0043587) taste bud development(GO:0061193) taste bud morphogenesis(GO:0061194) taste bud formation(GO:0061195) |
| 0.2 | 1.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.2 | 0.8 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.2 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.2 | 1.1 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.2 | 0.5 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 1.4 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.2 | 7.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 2.4 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.1 | 0.7 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.1 | 0.9 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 1.7 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 1.4 | GO:0010890 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.1 | 1.1 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 1.1 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.1 | 4.1 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 1.8 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 2.2 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.1 | 2.3 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.1 | 0.5 | GO:0060544 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 0.4 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.1 | 0.9 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.6 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 1.5 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.7 | GO:0044241 | lipid digestion(GO:0044241) |
| 0.1 | 1.0 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.1 | 0.7 | GO:0016559 | peroxisome fission(GO:0016559) mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 2.5 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 1.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 1.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 2.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.8 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 1.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 2.2 | GO:0098884 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 1.1 | GO:2000144 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.5 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) positive regulation of lamellipodium organization(GO:1902745) |
| 0.1 | 1.8 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.1 | 2.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 2.5 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.1 | 2.1 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 2.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 2.9 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 2.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 1.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 2.1 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 1.0 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 0.7 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 1.1 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.1 | 0.7 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.1 | 0.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.7 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.1 | 2.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 1.0 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 0.7 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 1.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 1.6 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) |
| 0.1 | 1.4 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.2 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 0.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.5 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.3 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 0.6 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 3.6 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.1 | 1.8 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.1 | 1.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.8 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.2 | GO:0046824 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.1 | 0.9 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 1.0 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.6 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.1 | 0.6 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.1 | 0.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.7 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 1.0 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.5 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.9 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.1 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 2.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.1 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.8 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 1.4 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 1.8 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.3 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.1 | 0.5 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 1.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.4 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 1.8 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 1.5 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 0.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.6 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.5 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 0.8 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 3.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.3 | GO:0051701 | interaction with host(GO:0051701) |
| 0.1 | 0.3 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 1.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.1 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.1 | 0.6 | GO:0071398 | cellular response to fatty acid(GO:0071398) |
| 0.1 | 3.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 2.0 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.1 | 1.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 2.8 | GO:0060840 | artery development(GO:0060840) |
| 0.1 | 0.6 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.1 | 0.4 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 0.3 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.1 | 1.7 | GO:0010906 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.1 | 0.2 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.1 | 1.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.5 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 2.6 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.1 | 1.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.2 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.1 | 1.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 2.7 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 1.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 3.9 | GO:0034976 | response to endoplasmic reticulum stress(GO:0034976) |
| 0.1 | 1.7 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.7 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.9 | GO:0071545 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 1.0 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 3.2 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 1.7 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 1.1 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 1.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 1.0 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.3 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 1.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.0 | 1.6 | GO:1902305 | regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 1.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 2.0 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.8 | GO:0009411 | response to UV(GO:0009411) |
| 0.0 | 0.8 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 0.7 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 2.5 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 1.1 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 1.1 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.9 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.0 | 1.2 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 1.3 | GO:0010107 | potassium ion import(GO:0010107) import across plasma membrane(GO:0098739) potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.3 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 1.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) medium-chain fatty acid catabolic process(GO:0051793) |
| 0.0 | 0.7 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.8 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 1.0 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.7 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.9 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.6 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.6 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 0.4 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.1 | GO:0016109 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.0 | 0.5 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 1.6 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.2 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.3 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 1.7 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.6 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) dendritic spine organization(GO:0097061) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.6 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 1.6 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.5 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 3.4 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.4 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.3 | GO:1900151 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of mRNA catabolic process(GO:0061013) positive regulation of mRNA catabolic process(GO:0061014) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.2 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.1 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.8 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.6 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0030237 | female sex determination(GO:0030237) |
| 0.0 | 0.7 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.5 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.7 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.0 | 3.8 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 1.1 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.2 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.0 | 3.6 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.6 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 0.4 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.4 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock(GO:0009649) entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.8 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.1 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0009446 | putrescine metabolic process(GO:0009445) putrescine biosynthetic process(GO:0009446) |
| 0.0 | 0.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 1.6 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.9 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 1.7 | GO:0016042 | lipid catabolic process(GO:0016042) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.6 | 2.4 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.6 | 8.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.5 | 2.5 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.4 | 1.7 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.4 | 1.3 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.4 | 2.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.4 | 2.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.4 | 1.1 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 1.7 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.3 | 1.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.3 | 3.9 | GO:0042627 | chylomicron(GO:0042627) |
| 0.3 | 4.8 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.3 | 1.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.3 | 0.8 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.2 | 0.7 | GO:1990745 | EARP complex(GO:1990745) |
| 0.2 | 1.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 1.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 20.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.2 | 1.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 1.7 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.2 | 4.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 2.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 3.4 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 0.8 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 3.0 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 3.1 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 1.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.5 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 5.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 2.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 1.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.6 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 1.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.0 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 3.6 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 1.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 8.4 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 11.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.5 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 3.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 3.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 11.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 1.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 1.2 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.8 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 1.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 2.5 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 1.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.5 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.1 | 0.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 3.6 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 1.5 | GO:0032590 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 2.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 1.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.9 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 4.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 1.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.4 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.5 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.8 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 2.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 3.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 2.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.1 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.8 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 6.8 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 7.1 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 2.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 2.0 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 2.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.3 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 2.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 14.0 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 4.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.7 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.8 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.0 | GO:0000786 | nucleosome(GO:0000786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 1.3 | 3.9 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 1.3 | 3.9 | GO:0042806 | fucose binding(GO:0042806) |
| 1.1 | 3.4 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 1.1 | 11.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.9 | 11.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.8 | 2.3 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) |
| 0.7 | 2.1 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.7 | 4.9 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.7 | 2.7 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.7 | 2.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.6 | 1.9 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.6 | 1.9 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.6 | 2.6 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.6 | 3.7 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.6 | 2.4 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.5 | 1.6 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.5 | 2.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.5 | 2.1 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.5 | 3.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.5 | 3.4 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.4 | 1.3 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.4 | 2.7 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.4 | 1.3 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.4 | 2.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.4 | 1.3 | GO:0001729 | ceramide kinase activity(GO:0001729) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.4 | 4.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 1.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.4 | 2.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.4 | 5.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.4 | 1.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.3 | 1.4 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.3 | 1.0 | GO:0034246 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.3 | 2.0 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.3 | 1.9 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.3 | 1.3 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.3 | 1.6 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.3 | 1.6 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.3 | 1.6 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.3 | 0.9 | GO:0052833 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.3 | 1.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.3 | 0.9 | GO:0000035 | acyl binding(GO:0000035) |
| 0.3 | 0.9 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.3 | 7.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 3.0 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 3.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.3 | 1.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.3 | 5.5 | GO:0005344 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.3 | 1.8 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.3 | 8.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.3 | 2.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 2.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 4.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 2.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 2.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.2 | 1.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 3.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 0.7 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.2 | 1.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 3.7 | GO:0015145 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.2 | 2.6 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.2 | 6.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 1.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 1.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 1.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 1.8 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 0.8 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.2 | 1.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 1.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.2 | 1.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 5.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 1.3 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.2 | 1.4 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 2.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 2.5 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.2 | 0.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 1.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 0.5 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 0.7 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.2 | 1.0 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.2 | 0.7 | GO:0043531 | ADP binding(GO:0043531) |
| 0.2 | 1.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 1.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.2 | 2.5 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.2 | 1.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 1.6 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.2 | 1.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 1.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 0.5 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 1.9 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 0.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 1.9 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 0.6 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 0.6 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 1.6 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 3.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.0 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 2.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.9 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 2.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 1.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 1.3 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 2.0 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 1.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 1.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 1.5 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.0 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.6 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 4.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 0.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 1.7 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 1.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.8 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 1.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.7 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 3.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 2.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 34.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.4 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 3.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 2.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 3.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 2.0 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.3 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.1 | 4.8 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.1 | 3.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 2.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.8 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 1.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.6 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 1.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.6 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 1.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.3 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.5 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.1 | 0.6 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 2.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.0 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 0.7 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 1.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 2.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.1 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 11.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.7 | GO:0019869 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 1.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 4.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 2.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.5 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.2 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 1.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 2.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0031779 | melanocortin receptor binding(GO:0031779) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.5 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.5 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.9 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 1.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 2.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 1.0 | GO:0015278 | calcium-release channel activity(GO:0015278) |
| 0.0 | 0.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.3 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.0 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 2.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.9 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 2.1 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 3.5 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.5 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.8 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 1.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 2.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 1.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.7 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.1 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 1.3 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.5 | GO:0000149 | SNARE binding(GO:0000149) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.2 | 4.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 1.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 7.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 3.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 5.7 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 3.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 1.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 2.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 5.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 0.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 2.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 0.6 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.5 | 9.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.5 | 2.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.4 | 3.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.4 | 3.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 2.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.3 | 4.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 2.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 6.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 2.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 5.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 3.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 1.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 1.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 1.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 1.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.3 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 1.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 2.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 2.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 6.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 4.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 0.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.0 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 1.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 1.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.7 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 0.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 0.8 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 2.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 1.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 4.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 2.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 2.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.2 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 2.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.1 | REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | Genes involved in Signaling by TGF-beta Receptor Complex |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.9 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |