Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
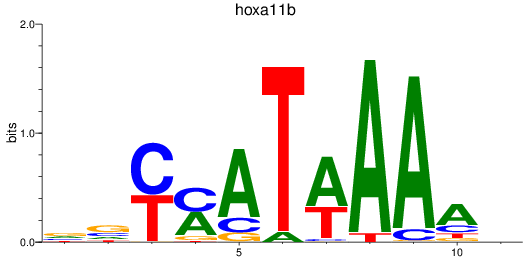
Results for hoxa11b
Z-value: 1.18
Transcription factors associated with hoxa11b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa11b
|
ENSDARG00000007009 | homeobox A11b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa11b | dr11_v1_chr16_+_20904754_20904754 | -0.38 | 2.0e-04 | Click! |
Activity profile of hoxa11b motif
Sorted Z-values of hoxa11b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_36585399 | 8.58 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr11_-_3343463 | 6.38 |
ENSDART00000066177
|
tuba2
|
tubulin, alpha 2 |
| chr8_-_44904723 | 6.15 |
ENSDART00000040804
|
praf2
|
PRA1 domain family, member 2 |
| chr10_-_24371312 | 5.88 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr3_+_46763745 | 5.68 |
ENSDART00000185437
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr24_-_6158933 | 5.51 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr20_+_29565906 | 5.42 |
ENSDART00000062383
|
ywhaqa
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide a |
| chr10_+_7029664 | 5.15 |
ENSDART00000166206
|
psd3l
|
pleckstrin and Sec7 domain containing 3, like |
| chr6_+_40661703 | 5.11 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr5_+_24245682 | 4.97 |
ENSDART00000049003
|
atp6v1aa
|
ATPase H+ transporting V1 subunit Aa |
| chr18_-_14677936 | 4.87 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr2_+_12349870 | 4.80 |
ENSDART00000134083
|
arhgap21b
|
Rho GTPase activating protein 21b |
| chr14_-_33297287 | 4.76 |
ENSDART00000045555
ENSDART00000138294 ENSDART00000075056 |
rab41
|
RAB41, member RAS oncogene family |
| chr3_+_46764022 | 4.67 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr8_+_36942262 | 4.60 |
ENSDART00000188173
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr6_-_42388608 | 4.58 |
ENSDART00000049425
|
sec61a1l
|
Sec61 translocon alpha 1 subunit, like |
| chr25_-_19433244 | 4.22 |
ENSDART00000154778
|
map1ab
|
microtubule-associated protein 1Ab |
| chr2_-_55298075 | 4.17 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr6_-_60147517 | 4.15 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr15_+_19324697 | 4.15 |
ENSDART00000022015
|
vps26b
|
VPS26 retromer complex component B |
| chr15_+_16387088 | 4.13 |
ENSDART00000101789
|
flot2b
|
flotillin 2b |
| chr2_+_35595454 | 4.03 |
ENSDART00000098734
|
cacybp
|
calcyclin binding protein |
| chr7_-_38634845 | 3.99 |
ENSDART00000173861
|
c1qtnf4
|
C1q and TNF related 4 |
| chr12_-_20616160 | 3.73 |
ENSDART00000105362
|
snx11
|
sorting nexin 11 |
| chr21_-_32781612 | 3.70 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr25_+_4750972 | 3.65 |
ENSDART00000168903
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr11_-_19775182 | 3.61 |
ENSDART00000037894
|
namptb
|
nicotinamide phosphoribosyltransferase b |
| chr11_-_10798021 | 3.59 |
ENSDART00000167112
ENSDART00000179725 ENSDART00000091923 ENSDART00000185825 |
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr3_+_24190207 | 3.58 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr25_-_27722309 | 3.56 |
ENSDART00000148121
|
zgc:153935
|
zgc:153935 |
| chr2_-_32738535 | 3.56 |
ENSDART00000135293
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr2_+_31948352 | 3.55 |
ENSDART00000192611
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr9_+_38163876 | 3.49 |
ENSDART00000137955
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr24_+_17007407 | 3.47 |
ENSDART00000110652
|
zfx
|
zinc finger protein, X-linked |
| chr1_-_20593778 | 3.46 |
ENSDART00000124770
|
ugt8
|
UDP glycosyltransferase 8 |
| chr10_+_25947946 | 3.46 |
ENSDART00000064393
|
ufm1
|
ubiquitin-fold modifier 1 |
| chr7_+_25059845 | 3.42 |
ENSDART00000077215
|
ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr23_-_43718067 | 3.41 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr18_+_31056645 | 3.39 |
ENSDART00000159316
|
mvda
|
mevalonate (diphospho) decarboxylase a |
| chr10_+_6013076 | 3.35 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr14_+_30795559 | 3.34 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr8_+_21229718 | 3.33 |
ENSDART00000100222
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr25_+_3994823 | 3.30 |
ENSDART00000154020
|
eps8l2
|
EPS8 like 2 |
| chr6_+_39493864 | 3.27 |
ENSDART00000086263
|
mettl7a
|
methyltransferase like 7A |
| chr13_-_25745089 | 3.24 |
ENSDART00000189333
ENSDART00000147420 |
sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr19_-_617246 | 3.23 |
ENSDART00000062551
|
cyp51
|
cytochrome P450, family 51 |
| chr16_+_7380463 | 3.19 |
ENSDART00000029727
ENSDART00000149086 |
atg5
|
ATG5 autophagy related 5 homolog (S. cerevisiae) |
| chr12_-_17152139 | 3.18 |
ENSDART00000152478
|
stambpl1
|
STAM binding protein-like 1 |
| chr17_-_23727978 | 3.15 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr16_+_36126310 | 3.15 |
ENSDART00000166040
ENSDART00000189802 |
sh3bp5b
|
SH3-domain binding protein 5b (BTK-associated) |
| chr8_-_17064243 | 3.12 |
ENSDART00000185313
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr23_-_30431333 | 3.09 |
ENSDART00000146633
|
camta1a
|
calmodulin binding transcription activator 1a |
| chr7_-_26076970 | 3.08 |
ENSDART00000101120
|
zgc:92664
|
zgc:92664 |
| chr16_-_42965192 | 3.08 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr9_+_34425736 | 3.05 |
ENSDART00000135147
|
si:ch211-218d20.15
|
si:ch211-218d20.15 |
| chr3_+_32105175 | 3.05 |
ENSDART00000113789
|
zgc:198419
|
zgc:198419 |
| chr7_-_52334840 | 3.01 |
ENSDART00000174173
|
CR938716.1
|
|
| chr21_+_21279159 | 3.01 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr22_+_10713713 | 3.01 |
ENSDART00000122349
|
hiat1b
|
hippocampus abundant transcript 1b |
| chr14_+_14841685 | 3.00 |
ENSDART00000158291
ENSDART00000162039 |
slbp
|
stem-loop binding protein |
| chr12_+_16087077 | 3.00 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr7_-_30082931 | 2.95 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr2_-_6115688 | 2.95 |
ENSDART00000081663
|
prdx1
|
peroxiredoxin 1 |
| chr16_-_55028740 | 2.94 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr14_+_23970818 | 2.90 |
ENSDART00000123338
ENSDART00000124944 |
kif3a
|
kinesin family member 3A |
| chr4_+_16885854 | 2.88 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr11_+_41090440 | 2.84 |
ENSDART00000188287
|
phf13
|
PHD finger protein 13 |
| chr22_+_6293563 | 2.82 |
ENSDART00000063416
|
rnasel2
|
ribonuclease like 2 |
| chr19_-_47571456 | 2.80 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr7_+_73801377 | 2.80 |
ENSDART00000184051
|
si:ch73-252p3.1
|
si:ch73-252p3.1 |
| chr8_+_52637507 | 2.80 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr6_+_3693441 | 2.80 |
ENSDART00000065256
|
ppig
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr24_-_21172122 | 2.76 |
ENSDART00000154259
|
atp6v1ab
|
ATPase H+ transporting V1 subunit Ab |
| chr1_+_15226268 | 2.72 |
ENSDART00000109911
|
hgsnat
|
heparan-alpha-glucosaminide N-acetyltransferase |
| chr17_+_39741926 | 2.71 |
ENSDART00000154996
ENSDART00000154599 |
si:dkey-229e3.2
|
si:dkey-229e3.2 |
| chr14_+_36738069 | 2.70 |
ENSDART00000105590
|
tdo2a
|
tryptophan 2,3-dioxygenase a |
| chr1_+_4101741 | 2.69 |
ENSDART00000163793
|
slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr13_-_9525527 | 2.68 |
ENSDART00000190618
|
CR848040.5
|
|
| chr10_-_36406344 | 2.67 |
ENSDART00000161823
|
ubl3a
|
ubiquitin-like 3a |
| chr24_-_40009446 | 2.65 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr11_-_6265574 | 2.63 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr15_-_29109620 | 2.62 |
ENSDART00000141007
|
zgc:162698
|
zgc:162698 |
| chr2_-_31735142 | 2.61 |
ENSDART00000130903
|
ralyl
|
RALY RNA binding protein like |
| chr12_+_3022882 | 2.61 |
ENSDART00000122905
|
rac3b
|
Rac family small GTPase 3b |
| chr15_-_1843831 | 2.60 |
ENSDART00000156718
ENSDART00000154175 |
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr12_-_4249000 | 2.59 |
ENSDART00000059298
|
zgc:92313
|
zgc:92313 |
| chr13_-_33022372 | 2.58 |
ENSDART00000147165
|
rbm25a
|
RNA binding motif protein 25a |
| chr18_-_48983690 | 2.58 |
ENSDART00000182359
|
FO681288.3
|
|
| chr22_-_16377960 | 2.56 |
ENSDART00000168170
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr8_-_25120231 | 2.56 |
ENSDART00000147308
|
amigo1
|
adhesion molecule with Ig-like domain 1 |
| chr5_+_28398449 | 2.55 |
ENSDART00000165292
|
nsmfb
|
NMDA receptor synaptonuclear signaling and neuronal migration factor b |
| chr17_-_30521043 | 2.54 |
ENSDART00000087111
|
itsn2b
|
intersectin 2b |
| chr17_-_31719071 | 2.53 |
ENSDART00000136199
|
dtd2
|
D-tyrosyl-tRNA deacylase 2 |
| chr22_-_10440688 | 2.50 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr15_+_23799461 | 2.49 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr19_+_4916233 | 2.49 |
ENSDART00000159512
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr12_-_19346678 | 2.48 |
ENSDART00000044860
|
maff
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr22_+_724639 | 2.47 |
ENSDART00000105323
|
zgc:162255
|
zgc:162255 |
| chr16_-_21047872 | 2.46 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr19_-_30447611 | 2.45 |
ENSDART00000073705
ENSDART00000048977 ENSDART00000191237 |
abcf1
|
ATP-binding cassette, sub-family F (GCN20), member 1 |
| chr9_-_48214216 | 2.45 |
ENSDART00000012938
|
phgdh
|
phosphoglycerate dehydrogenase |
| chr13_+_43400443 | 2.45 |
ENSDART00000084321
|
dact2
|
dishevelled-binding antagonist of beta-catenin 2 |
| chr17_-_31611692 | 2.44 |
ENSDART00000141480
|
si:dkey-170l10.1
|
si:dkey-170l10.1 |
| chr25_-_27722614 | 2.43 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr16_+_7626535 | 2.43 |
ENSDART00000182670
ENSDART00000065514 ENSDART00000150212 |
stx12l
|
syntaxin 12, like |
| chr20_-_34028967 | 2.43 |
ENSDART00000153408
ENSDART00000033817 |
scyl3
|
SCY1-like, kinase-like 3 |
| chr5_-_4532516 | 2.40 |
ENSDART00000192398
|
cst14b.1
|
cystatin 14b, tandem duplicate 1 |
| chr19_-_31402429 | 2.39 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr6_+_45918981 | 2.39 |
ENSDART00000149642
|
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr21_-_2299002 | 2.39 |
ENSDART00000168712
|
si:ch73-299h12.6
|
si:ch73-299h12.6 |
| chr17_+_48126944 | 2.38 |
ENSDART00000103397
|
slc39a9
|
solute carrier family 39, member 9 |
| chr19_+_42061699 | 2.38 |
ENSDART00000125579
|
si:ch211-13c6.2
|
si:ch211-13c6.2 |
| chr3_-_40976288 | 2.37 |
ENSDART00000193553
|
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr2_-_59145027 | 2.36 |
ENSDART00000128320
|
FO834803.1
|
|
| chr2_-_21170517 | 2.36 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr14_+_33264303 | 2.32 |
ENSDART00000130680
ENSDART00000075187 |
pdzd11
|
PDZ domain containing 11 |
| chr10_-_25328814 | 2.31 |
ENSDART00000123820
|
tmem135
|
transmembrane protein 135 |
| chr25_+_35913614 | 2.31 |
ENSDART00000022437
|
gpia
|
glucose-6-phosphate isomerase a |
| chr17_+_23300827 | 2.30 |
ENSDART00000058745
|
zgc:165461
|
zgc:165461 |
| chr16_-_17586883 | 2.30 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr14_+_34966598 | 2.29 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr14_+_45565891 | 2.29 |
ENSDART00000133389
ENSDART00000025549 |
zgc:92249
|
zgc:92249 |
| chr1_-_625875 | 2.28 |
ENSDART00000167331
|
appa
|
amyloid beta (A4) precursor protein a |
| chr12_-_25887864 | 2.27 |
ENSDART00000152983
|
si:dkey-193p11.2
|
si:dkey-193p11.2 |
| chr3_+_24189804 | 2.27 |
ENSDART00000134723
|
prr15la
|
proline rich 15-like a |
| chr10_+_7593185 | 2.26 |
ENSDART00000162617
ENSDART00000162590 ENSDART00000171744 |
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr3_+_45368973 | 2.26 |
ENSDART00000187282
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr16_+_39146696 | 2.25 |
ENSDART00000121756
ENSDART00000084381 |
sybu
|
syntabulin (syntaxin-interacting) |
| chr4_+_27130412 | 2.25 |
ENSDART00000145083
|
brd1a
|
bromodomain containing 1a |
| chr22_+_8753092 | 2.25 |
ENSDART00000140720
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr3_+_62161184 | 2.24 |
ENSDART00000090370
ENSDART00000192665 |
noxo1a
|
NADPH oxidase organizer 1a |
| chr15_+_17321218 | 2.24 |
ENSDART00000143796
|
cltcb
|
clathrin, heavy chain b (Hc) |
| chr2_+_3823813 | 2.24 |
ENSDART00000103596
ENSDART00000161880 ENSDART00000185408 |
npc1
|
Niemann-Pick disease, type C1 |
| chr17_+_31221761 | 2.23 |
ENSDART00000155580
|
ccdc32
|
coiled-coil domain containing 32 |
| chr5_+_58687541 | 2.23 |
ENSDART00000083015
ENSDART00000181902 |
ccdc84
|
coiled-coil domain containing 84 |
| chr23_+_17354154 | 2.23 |
ENSDART00000155808
|
SRMS
|
zgc:194282 |
| chr15_+_43166511 | 2.22 |
ENSDART00000011737
|
flj13639
|
flj13639 |
| chr16_+_13883872 | 2.21 |
ENSDART00000101304
ENSDART00000136005 |
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr5_-_26765188 | 2.21 |
ENSDART00000029450
|
rnf181
|
ring finger protein 181 |
| chr10_-_36406562 | 2.20 |
ENSDART00000160468
|
ubl3a
|
ubiquitin-like 3a |
| chr2_+_10147029 | 2.18 |
ENSDART00000139064
ENSDART00000053426 ENSDART00000153678 |
pfn2l
|
profilin 2 like |
| chr5_+_1278092 | 2.17 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr6_-_29377092 | 2.16 |
ENSDART00000078665
|
tmem131
|
transmembrane protein 131 |
| chr2_+_58841181 | 2.16 |
ENSDART00000164102
|
cirbpa
|
cold inducible RNA binding protein a |
| chr22_-_20166660 | 2.16 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr13_+_28690355 | 2.16 |
ENSDART00000137475
ENSDART00000128246 |
polr1c
|
polymerase (RNA) I polypeptide C |
| chr20_-_31905968 | 2.15 |
ENSDART00000142806
|
stxbp5a
|
syntaxin binding protein 5a (tomosyn) |
| chr10_+_5234327 | 2.15 |
ENSDART00000133927
ENSDART00000063120 |
sptlc1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr13_-_36034582 | 2.15 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr18_+_21122818 | 2.15 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr21_-_22115136 | 2.14 |
ENSDART00000134715
ENSDART00000089246 ENSDART00000139789 |
elmod1
|
ELMO/CED-12 domain containing 1 |
| chr12_+_31638045 | 2.14 |
ENSDART00000184216
ENSDART00000183645 ENSDART00000153129 |
dnmbp
|
dynamin binding protein |
| chr15_+_42285643 | 2.13 |
ENSDART00000152731
|
scaf4b
|
SR-related CTD-associated factor 4b |
| chr10_-_8060573 | 2.12 |
ENSDART00000147104
ENSDART00000099030 |
si:ch211-251f6.6
|
si:ch211-251f6.6 |
| chr24_+_36840652 | 2.12 |
ENSDART00000088168
|
CABZ01055347.1
|
|
| chr7_+_67749251 | 2.12 |
ENSDART00000167562
|
dhx38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr5_-_1274794 | 2.11 |
ENSDART00000167337
ENSDART00000125247 |
ciz1a
|
cdkn1a interacting zinc finger protein 1a |
| chr5_+_50913034 | 2.11 |
ENSDART00000149787
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr6_-_18400548 | 2.11 |
ENSDART00000179797
ENSDART00000164891 |
trim25
|
tripartite motif containing 25 |
| chr2_+_58008980 | 2.11 |
ENSDART00000171264
|
si:ch211-155e24.3
|
si:ch211-155e24.3 |
| chr18_+_5875268 | 2.11 |
ENSDART00000177784
ENSDART00000122009 |
wdr59
|
WD repeat domain 59 |
| chr5_-_45877387 | 2.10 |
ENSDART00000183714
ENSDART00000041503 |
slc4a4a
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4a |
| chr16_+_9400661 | 2.09 |
ENSDART00000146174
|
ice1
|
KIAA0947-like (H. sapiens) |
| chr25_-_11378623 | 2.08 |
ENSDART00000166586
|
enc2
|
ectodermal-neural cortex 2 |
| chr23_+_20518504 | 2.08 |
ENSDART00000114246
|
adnpb
|
activity-dependent neuroprotector homeobox b |
| chr15_-_34418525 | 2.08 |
ENSDART00000147582
|
agmo
|
alkylglycerol monooxygenase |
| chr14_+_49135264 | 2.07 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr14_+_23717165 | 2.07 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr1_+_27868294 | 2.07 |
ENSDART00000165332
|
dnajb14
|
DnaJ (Hsp40) homolog, subfamily B, member 14 |
| chr16_-_42390640 | 2.06 |
ENSDART00000193214
ENSDART00000102305 |
cspg5a
|
chondroitin sulfate proteoglycan 5a |
| chr20_+_32497260 | 2.06 |
ENSDART00000145175
ENSDART00000132921 |
si:ch73-257c13.2
|
si:ch73-257c13.2 |
| chr14_-_33481428 | 2.05 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr8_+_3820134 | 2.05 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr22_+_18156000 | 2.05 |
ENSDART00000143483
ENSDART00000136133 |
nr2c2ap
|
nuclear receptor 2C2-associated protein |
| chr5_-_28767573 | 2.04 |
ENSDART00000158299
ENSDART00000043466 |
traf2a
|
Tnf receptor-associated factor 2a |
| chr16_+_43401005 | 2.04 |
ENSDART00000110994
|
sqlea
|
squalene epoxidase a |
| chr16_+_28578352 | 2.02 |
ENSDART00000149306
|
nmt2
|
N-myristoyltransferase 2 |
| chr7_-_30779575 | 2.02 |
ENSDART00000004782
|
mphosph10
|
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr13_+_37656278 | 2.01 |
ENSDART00000193251
|
phf3
|
PHD finger protein 3 |
| chr18_+_30028637 | 2.01 |
ENSDART00000139750
|
si:ch211-220f16.1
|
si:ch211-220f16.1 |
| chr7_+_42935126 | 2.00 |
ENSDART00000157747
|
BX284696.1
|
|
| chr14_+_45566074 | 2.00 |
ENSDART00000133477
ENSDART00000185899 |
zgc:92249
|
zgc:92249 |
| chr3_-_26184018 | 1.99 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr19_-_47571797 | 1.98 |
ENSDART00000166180
ENSDART00000168134 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr5_-_51209101 | 1.97 |
ENSDART00000172006
ENSDART00000142455 |
sdccag3
|
serologically defined colon cancer antigen 3 |
| chr12_+_3262564 | 1.96 |
ENSDART00000184264
|
tmem101
|
transmembrane protein 101 |
| chr3_-_19495814 | 1.96 |
ENSDART00000162248
|
CU571315.3
|
|
| chr20_-_29499363 | 1.95 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr5_-_36597612 | 1.95 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr6_+_7444899 | 1.95 |
ENSDART00000053775
|
arf3b
|
ADP-ribosylation factor 3b |
| chr3_-_21037840 | 1.94 |
ENSDART00000002393
|
rundc3aa
|
RUN domain containing 3Aa |
| chr19_+_1370504 | 1.94 |
ENSDART00000158946
|
dgat1a
|
diacylglycerol O-acyltransferase 1a |
| chr17_-_10043273 | 1.94 |
ENSDART00000156078
|
baz1a
|
bromodomain adjacent to zinc finger domain, 1A |
| chr16_+_46111849 | 1.94 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr22_+_2207502 | 1.93 |
ENSDART00000169162
|
si:dkeyp-79b7.12
|
si:dkeyp-79b7.12 |
| chr17_+_10501647 | 1.93 |
ENSDART00000140391
|
tyro3
|
TYRO3 protein tyrosine kinase |
| chr8_-_10961991 | 1.93 |
ENSDART00000139603
|
trim33
|
tripartite motif containing 33 |
| chr3_-_61387273 | 1.93 |
ENSDART00000156479
|
znf1143
|
zinc finger protein 1143 |
| chr3_-_32337653 | 1.92 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr15_-_1844048 | 1.92 |
ENSDART00000102410
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa11b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.6 | 4.8 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 1.1 | 3.3 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 1.1 | 3.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 1.0 | 4.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.9 | 2.6 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.8 | 3.4 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.8 | 2.5 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.7 | 2.1 | GO:0039529 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.7 | 3.5 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.7 | 4.6 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.6 | 5.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.6 | 1.9 | GO:0071047 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.6 | 2.5 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.6 | 4.3 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.6 | 6.7 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.6 | 1.8 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.6 | 1.7 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.6 | 2.3 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.6 | 2.3 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.6 | 3.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.5 | 2.7 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.5 | 1.6 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.5 | 4.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.5 | 11.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.5 | 4.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.5 | 1.9 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.5 | 2.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.4 | 2.2 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.4 | 3.0 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.4 | 1.3 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.4 | 1.3 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.4 | 2.1 | GO:0030910 | olfactory placode formation(GO:0030910) |
| 0.4 | 2.5 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.4 | 2.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.4 | 2.5 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.4 | 1.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.4 | 4.8 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.4 | 1.9 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.4 | 1.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.4 | 3.4 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.4 | 1.9 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.4 | 1.5 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.4 | 2.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.4 | 1.8 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.4 | 2.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.4 | 2.5 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.3 | 1.0 | GO:1902230 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.3 | 2.7 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.3 | 2.0 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.3 | 7.5 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.3 | 1.9 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.3 | 1.0 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.3 | 1.3 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.3 | 1.0 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.3 | 1.3 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.3 | 0.9 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.3 | 1.9 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.3 | 1.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.3 | 2.5 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.3 | 1.5 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.3 | 1.8 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.3 | 2.9 | GO:0019430 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.3 | 0.6 | GO:0098586 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.3 | 1.1 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.3 | 3.4 | GO:0006415 | translational termination(GO:0006415) |
| 0.3 | 1.0 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.3 | 1.3 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.3 | 0.8 | GO:0001783 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.3 | 1.8 | GO:0015809 | arginine transport(GO:0015809) |
| 0.3 | 2.5 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.3 | 0.8 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.3 | 1.0 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.3 | 1.3 | GO:0045056 | transcytosis(GO:0045056) |
| 0.2 | 5.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.2 | 5.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 1.5 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.2 | 1.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 14.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.2 | 5.3 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.2 | 0.7 | GO:0060148 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.2 | 6.1 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.2 | 1.6 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.2 | 1.2 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.2 | 1.4 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 1.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 0.7 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.2 | 9.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 2.9 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 1.5 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.2 | 1.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 2.1 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.2 | 0.8 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.2 | 3.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.2 | 3.0 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.2 | 1.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.2 | 1.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.2 | 0.7 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 0.7 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.2 | 0.9 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.2 | 0.9 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.2 | 0.5 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.2 | 2.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 0.9 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.2 | 0.5 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.2 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 2.3 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.2 | 1.9 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 1.0 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 3.2 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.2 | 0.8 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 2.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 | 1.0 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.2 | 0.8 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.2 | 0.8 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.2 | 0.8 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 0.7 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.2 | 1.8 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.2 | 2.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.2 | 9.5 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.2 | 2.0 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.2 | 3.5 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.2 | 2.6 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.2 | 1.7 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.2 | 0.8 | GO:0032370 | positive regulation of lipid transport(GO:0032370) regulation of sterol transport(GO:0032371) regulation of cholesterol transport(GO:0032374) |
| 0.2 | 2.6 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.2 | 3.0 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 3.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.9 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.6 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.3 | GO:1900180 | regulation of protein localization to nucleus(GO:1900180) |
| 0.1 | 3.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 3.3 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 1.7 | GO:0031577 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.1 | 1.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 1.5 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 3.5 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.1 | 0.8 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 1.1 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.7 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.9 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 2.6 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.1 | 1.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.8 | GO:0097065 | anterior head development(GO:0097065) |
| 0.1 | 0.6 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.1 | 3.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 0.9 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 0.1 | 2.2 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 4.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.9 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.9 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 0.7 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 2.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 3.5 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.1 | 0.4 | GO:0002544 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 0.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 3.7 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.1 | 1.8 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 1.3 | GO:0046883 | regulation of hormone secretion(GO:0046883) |
| 0.1 | 1.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.3 | GO:0097623 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 0.1 | 2.7 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 0.4 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.1 | 1.3 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 1.2 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.7 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 0.8 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 1.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) synaptic vesicle budding(GO:0070142) |
| 0.1 | 1.0 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.2 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.1 | 0.6 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.6 | GO:1902915 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 6.7 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.1 | 2.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 1.2 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.1 | 2.6 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 1.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.2 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.1 | 2.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.6 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.1 | 0.4 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 2.0 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.1 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 2.2 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 1.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 1.4 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.1 | 2.8 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 1.4 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 1.2 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.9 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.9 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 0.8 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 1.2 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.1 | 0.6 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.1 | 0.2 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.1 | 1.8 | GO:0000302 | response to reactive oxygen species(GO:0000302) |
| 0.1 | 2.5 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.1 | 1.5 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.1 | 6.3 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 1.9 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.1 | 0.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.8 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 1.6 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 1.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.3 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 1.8 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 0.5 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.1 | 0.5 | GO:0034340 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 1.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 1.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 1.4 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.1 | 0.7 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 1.8 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 0.6 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 1.1 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 0.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 1.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 1.9 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.1 | 0.2 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 | 1.0 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 1.6 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.2 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 | 2.3 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.1 | 0.7 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 0.3 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.9 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.9 | GO:0000959 | mitochondrial RNA metabolic process(GO:0000959) |
| 0.0 | 1.2 | GO:0048920 | neuromast primordium migration(GO:0048883) posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 2.3 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 1.2 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 1.7 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.7 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.8 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.9 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.7 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.5 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 1.0 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.9 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 1.0 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.8 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 1.4 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 0.9 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 3.9 | GO:0016072 | rRNA processing(GO:0006364) rRNA metabolic process(GO:0016072) |
| 0.0 | 3.6 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 1.5 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 10.3 | GO:0000375 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.5 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 1.4 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 0.7 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 1.0 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 3.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 2.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.4 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 1.8 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 1.3 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.3 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 1.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.8 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 1.6 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.8 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 19.2 | GO:0015031 | protein transport(GO:0015031) |
| 0.0 | 3.0 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 1.4 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 2.8 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 1.2 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 3.7 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 2.0 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 1.5 | GO:0044782 | cilium organization(GO:0044782) |
| 0.0 | 0.6 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 3.4 | GO:0006399 | tRNA metabolic process(GO:0006399) |
| 0.0 | 0.7 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.9 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.8 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.7 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 2.3 | GO:0045786 | negative regulation of cell cycle(GO:0045786) |
| 0.0 | 0.1 | GO:0051383 | kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.0 | 0.7 | GO:0071600 | otic vesicle morphogenesis(GO:0071600) |
| 0.0 | 0.8 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.7 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 1.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 2.7 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 4.4 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 0.6 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 1.1 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 0.1 | GO:0030237 | female sex determination(GO:0030237) male sex determination(GO:0030238) |
| 0.0 | 0.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 3.3 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 1.9 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.9 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 1.5 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 2.6 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 1.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.5 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.1 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.0 | 0.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.0 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.4 | GO:1903038 | negative regulation of homotypic cell-cell adhesion(GO:0034111) negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 5.9 | GO:1902679 | negative regulation of transcription, DNA-templated(GO:0045892) negative regulation of RNA biosynthetic process(GO:1902679) negative regulation of nucleic acid-templated transcription(GO:1903507) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.5 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.1 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.9 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.7 | GO:0006338 | chromatin remodeling(GO:0006338) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.3 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 1.2 | 3.5 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 1.0 | 4.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.8 | 4.6 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.7 | 2.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.7 | 5.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.7 | 2.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.7 | 3.9 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.6 | 5.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.6 | 5.0 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.5 | 5.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.5 | 3.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.5 | 1.9 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.5 | 7.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.4 | 2.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.4 | 1.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.4 | 1.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.4 | 1.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.4 | 1.6 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.4 | 1.5 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.3 | 1.6 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.3 | 4.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 1.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 5.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.2 | 1.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 1.2 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.2 | 0.9 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 1.6 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.2 | 7.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 1.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 0.8 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 1.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 3.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 7.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 0.8 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.2 | 1.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 1.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.2 | 2.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 1.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 1.9 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 1.2 | GO:0001650 | fibrillar center(GO:0001650) multimeric ribonuclease P complex(GO:0030681) |
| 0.2 | 3.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 0.6 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.2 | 3.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 1.1 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 2.8 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 1.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 3.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 1.0 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 2.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 5.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 1.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 4.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 2.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 6.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.5 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.4 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 1.8 | GO:1990752 | microtubule end(GO:1990752) |
| 0.1 | 0.7 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 6.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 1.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.1 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 1.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 12.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 0.4 | GO:0043514 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.1 | 0.8 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 1.8 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.1 | 2.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.7 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 1.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 3.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.0 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 0.2 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.1 | 1.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.3 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 5.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.0 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 2.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 4.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 5.9 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 1.7 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 6.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 1.2 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 7.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 0.9 | GO:0005801 | cis-Golgi network(GO:0005801) TRAPP complex(GO:0030008) |
| 0.1 | 0.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.1 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 5.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 1.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 4.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 5.6 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.8 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.8 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 3.9 | GO:0031984 | trans-Golgi network(GO:0005802) organelle subcompartment(GO:0031984) |
| 0.0 | 2.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 5.8 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 2.1 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.7 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 7.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.8 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 1.0 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 4.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 6.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 1.1 | 3.3 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 1.0 | 4.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 1.0 | 6.7 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.9 | 2.7 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.8 | 2.5 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.8 | 5.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.8 | 7.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.8 | 3.2 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.8 | 2.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.8 | 3.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.8 | 2.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.7 | 7.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.7 | 3.4 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.6 | 5.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.6 | 1.9 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.6 | 2.3 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.5 | 2.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.5 | 1.5 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.5 | 1.4 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.5 | 1.9 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.5 | 1.4 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.4 | 3.9 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.4 | 1.3 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.4 | 3.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.4 | 1.3 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.4 | 1.7 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.4 | 5.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.4 | 3.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 1.9 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.4 | 5.0 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.4 | 4.6 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.4 | 5.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.4 | 5.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.4 | 1.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.3 | 3.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.3 | 1.4 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.3 | 1.0 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.3 | 1.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.3 | 1.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.3 | 2.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.3 | 2.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 1.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.3 | 1.5 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.3 | 2.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 1.2 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.3 | 1.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.3 | 1.6 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.3 | 3.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.3 | 1.0 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.3 | 2.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.3 | 1.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.3 | 0.8 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 2.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 1.9 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 0.9 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.2 | 1.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 0.6 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.2 | 6.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 0.6 | GO:0004788 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.2 | 0.8 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.2 | 1.0 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.2 | 3.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.4 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 3.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 1.6 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.2 | 1.9 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.2 | 0.9 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 3.0 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.2 | 2.0 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 2.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 0.9 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.2 | 2.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 2.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 3.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 2.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.2 | 1.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 0.9 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.2 | 0.3 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.2 | 0.9 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 3.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 0.5 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.1 | 0.7 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.9 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.1 | 1.8 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 5.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 2.7 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 3.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 3.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 2.2 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.1 | 1.4 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 6.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 3.2 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.1 | 1.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 2.0 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 3.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 2.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 3.0 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.6 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.9 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 1.4 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.1 | 0.8 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.1 | 3.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.7 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.7 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 1.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 2.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.3 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.1 | 0.4 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.1 | 1.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.6 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.3 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.1 | 1.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 7.3 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 0.8 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 1.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 1.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 3.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 5.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 7.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.6 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.1 | 0.7 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.7 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 2.2 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 3.2 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.1 | 1.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 2.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.2 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.7 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 4.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 1.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.7 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 1.8 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.7 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 3.9 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 0.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 1.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.9 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 1.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 2.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.1 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.1 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.3 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 1.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 3.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.1 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.9 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 1.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 1.3 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 1.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 1.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 5.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 14.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 8.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 5.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.5 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 1.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 1.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 8.6 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 7.0 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 7.2 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 12.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 2.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 3.1 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 3.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.0 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 2.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.6 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 2.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.2 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 7.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 3.1 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 4.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.8 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 2.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.9 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 1.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 3.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 2.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 7.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 6.4 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 3.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 3.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 3.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 3.3 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 4.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 5.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.9 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 3.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 3.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.6 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 1.0 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 10.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.7 | 9.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.5 | 7.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.4 | 5.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.3 | 3.0 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.3 | 4.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 3.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 6.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 3.0 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.3 | 1.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.3 | 3.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.3 | 7.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.3 | 3.6 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.3 | 1.8 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 7.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 2.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 0.6 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.2 | 5.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.2 | 1.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.2 | 2.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 3.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 1.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.3 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 1.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 5.6 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.1 | 1.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 1.0 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 4.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 1.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 0.8 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 3.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 4.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.8 | REACTOME MITOTIC G2 G2 M PHASES | Genes involved in Mitotic G2-G2/M phases |
| 0.1 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 3.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 2.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 3.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.0 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.7 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 2.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 3.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.6 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |