Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
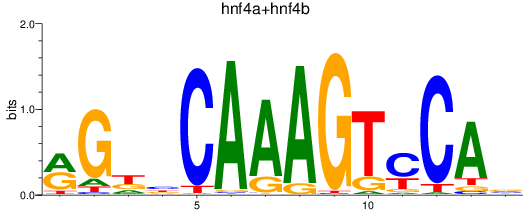
Results for hnf4a+hnf4b
Z-value: 4.57
Transcription factors associated with hnf4a+hnf4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hnf4a
|
ENSDARG00000021494 | hepatocyte nuclear factor 4, alpha |
|
hnf4b
|
ENSDARG00000104742 | hepatic nuclear factor 4, beta |
|
hnf4a
|
ENSDARG00000112985 | hepatocyte nuclear factor 4, alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hnf4a | dr11_v1_chr23_+_25832689_25832689 | 0.90 | 4.9e-34 | Click! |
| CABZ01057488.1 | dr11_v1_chr7_+_69019851_69019851 | 0.88 | 6.9e-31 | Click! |
Activity profile of hnf4a+hnf4b motif
Sorted Z-values of hnf4a+hnf4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_23913943 | 156.24 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr20_+_1412193 | 139.48 |
ENSDART00000064419
|
leg1.1
|
liver-enriched gene 1, tandem duplicate 1 |
| chr20_+_1398564 | 115.45 |
ENSDART00000002242
|
leg1.2
|
liver-enriched gene 1, tandem duplicate 2 |
| chr1_+_17900306 | 109.48 |
ENSDART00000089480
|
cyp4v8
|
cytochrome P450, family 4, subfamily V, polypeptide 8 |
| chr15_-_26552393 | 101.89 |
ENSDART00000150152
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr1_+_53374454 | 88.09 |
ENSDART00000038807
|
ucp1
|
uncoupling protein 1 |
| chr15_-_26552652 | 87.13 |
ENSDART00000152336
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr16_+_23923746 | 86.54 |
ENSDART00000137962
|
si:dkey-7f3.14
|
si:dkey-7f3.14 |
| chr17_-_30702411 | 86.30 |
ENSDART00000114358
|
zgc:194392
|
zgc:194392 |
| chr19_+_9277327 | 85.66 |
ENSDART00000104623
ENSDART00000151164 |
si:rp71-15k1.1
|
si:rp71-15k1.1 |
| chr2_-_37960688 | 83.10 |
ENSDART00000055565
|
cbln14
|
cerebellin 14 |
| chr7_+_25036188 | 78.50 |
ENSDART00000163957
ENSDART00000169749 |
sb:cb1058
|
sb:cb1058 |
| chr2_-_37956768 | 72.62 |
ENSDART00000034595
|
cbln10
|
cerebellin 10 |
| chr10_+_17026870 | 72.41 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr13_-_22843562 | 71.98 |
ENSDART00000142738
|
pbld
|
phenazine biosynthesis like protein domain containing |
| chr8_-_30791266 | 70.51 |
ENSDART00000062220
|
gstt1a
|
glutathione S-transferase theta 1a |
| chr22_-_20126230 | 68.70 |
ENSDART00000138688
|
creb3l3a
|
cAMP responsive element binding protein 3-like 3a |
| chr9_-_9982696 | 68.03 |
ENSDART00000192548
ENSDART00000125852 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr18_+_17611627 | 67.91 |
ENSDART00000046891
|
cetp
|
cholesteryl ester transfer protein, plasma |
| chr8_+_30699429 | 66.82 |
ENSDART00000005345
|
upb1
|
ureidopropionase, beta |
| chr20_-_40754794 | 64.67 |
ENSDART00000187251
|
cx32.3
|
connexin 32.3 |
| chr8_+_1766206 | 64.35 |
ENSDART00000021820
|
serpind1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr14_+_21107032 | 63.72 |
ENSDART00000138319
ENSDART00000139103 ENSDART00000184735 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr22_+_37888249 | 63.64 |
ENSDART00000076082
|
fetub
|
fetuin B |
| chr16_+_23947196 | 61.79 |
ENSDART00000103190
ENSDART00000132961 ENSDART00000147690 ENSDART00000142168 |
apoa4b.2
|
apolipoprotein A-IV b, tandem duplicate 2 |
| chr21_-_27443995 | 61.58 |
ENSDART00000003508
|
bfb
|
complement component bfb |
| chr16_-_17197546 | 61.49 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr25_-_20049449 | 60.77 |
ENSDART00000104315
|
zgc:136858
|
zgc:136858 |
| chr16_+_23924040 | 60.28 |
ENSDART00000161124
|
si:dkey-7f3.14
|
si:dkey-7f3.14 |
| chr8_-_37263524 | 59.74 |
ENSDART00000061327
|
rh50
|
Rh50-like protein |
| chr12_-_6172154 | 58.85 |
ENSDART00000185434
|
a1cf
|
apobec1 complementation factor |
| chr10_-_322769 | 58.66 |
ENSDART00000165244
|
akt2l
|
v-akt murine thymoma viral oncogene homolog 2, like |
| chr3_-_45848257 | 57.34 |
ENSDART00000147198
|
igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr17_+_20173882 | 57.19 |
ENSDART00000155379
|
si:ch211-248a14.8
|
si:ch211-248a14.8 |
| chr18_+_40462445 | 54.91 |
ENSDART00000087645
|
ugt5c2
|
UDP glucuronosyltransferase 5 family, polypeptide C2 |
| chr16_-_17200120 | 54.79 |
ENSDART00000147739
|
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr22_-_24818066 | 51.46 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr10_+_26747755 | 50.37 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr7_+_52135791 | 49.40 |
ENSDART00000098705
|
cyp2x12
|
cytochrome P450, family 2, subfamily X, polypeptide 12 |
| chr8_+_39802506 | 48.47 |
ENSDART00000018862
|
hnf1a
|
HNF1 homeobox a |
| chr2_+_22659787 | 47.40 |
ENSDART00000043956
|
zgc:161973
|
zgc:161973 |
| chr2_+_3533458 | 47.21 |
ENSDART00000133007
|
gpt2l
|
glutamic pyruvate transaminase (alanine aminotransferase) 2, like |
| chr2_-_37967368 | 45.87 |
ENSDART00000050345
|
cbln9
|
cerebellin 9 |
| chr3_+_16762483 | 45.19 |
ENSDART00000132732
|
tmem86b
|
transmembrane protein 86B |
| chr25_-_26758253 | 44.69 |
ENSDART00000123004
|
si:dkeyp-73b11.8
|
si:dkeyp-73b11.8 |
| chr6_-_1768724 | 43.70 |
ENSDART00000162488
ENSDART00000163613 |
zgc:158417
|
zgc:158417 |
| chr13_-_40726865 | 42.79 |
ENSDART00000099847
|
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr16_+_23921610 | 42.28 |
ENSDART00000143855
|
apoa4b.3
|
apolipoprotein A-IV b, tandem duplicate 3 |
| chr18_+_18861359 | 42.27 |
ENSDART00000144605
|
pllp
|
plasmolipin |
| chr23_-_27702561 | 42.18 |
ENSDART00000053876
|
dnajc22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr17_+_20174044 | 41.90 |
ENSDART00000156028
|
si:ch211-248a14.8
|
si:ch211-248a14.8 |
| chr13_+_50375800 | 40.35 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr5_-_69940868 | 38.84 |
ENSDART00000185924
ENSDART00000097357 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr16_+_32029090 | 37.75 |
ENSDART00000041054
|
tmc4
|
transmembrane channel-like 4 |
| chr24_-_17047918 | 37.45 |
ENSDART00000020204
|
msrb2
|
methionine sulfoxide reductase B2 |
| chr2_-_24407933 | 35.15 |
ENSDART00000088584
|
si:dkey-208k22.6
|
si:dkey-208k22.6 |
| chr2_+_3533117 | 35.09 |
ENSDART00000132572
|
gpt2l
|
glutamic pyruvate transaminase (alanine aminotransferase) 2, like |
| chr6_+_27624023 | 34.57 |
ENSDART00000147789
|
slco2a1
|
solute carrier organic anion transporter family, member 2A1 |
| chr5_-_62317496 | 33.63 |
ENSDART00000180089
|
zgc:85789
|
zgc:85789 |
| chr16_+_23921777 | 33.08 |
ENSDART00000163213
|
apoa4b.3
|
apolipoprotein A-IV b, tandem duplicate 3 |
| chr17_-_49407091 | 32.35 |
ENSDART00000021950
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr20_+_54304800 | 30.77 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr23_-_24488696 | 30.66 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr16_-_25607266 | 30.57 |
ENSDART00000192602
|
zgc:110410
|
zgc:110410 |
| chr23_-_44786844 | 30.33 |
ENSDART00000148669
|
si:ch73-269m23.5
|
si:ch73-269m23.5 |
| chr8_-_65189 | 29.94 |
ENSDART00000168412
|
hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr5_-_3839285 | 29.01 |
ENSDART00000122292
|
mlxipl
|
MLX interacting protein like |
| chr13_+_8696825 | 28.99 |
ENSDART00000109059
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr24_-_36727922 | 28.42 |
ENSDART00000135142
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr3_+_12744083 | 28.07 |
ENSDART00000158554
ENSDART00000169545 |
cyp2k21
|
cytochrome P450, family 2, subfamily k, polypeptide 21 |
| chr17_-_2584423 | 28.06 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr15_-_24178893 | 27.44 |
ENSDART00000077980
|
pipox
|
pipecolic acid oxidase |
| chr19_+_31532043 | 27.33 |
ENSDART00000136289
|
tmem64
|
transmembrane protein 64 |
| chr5_-_3885727 | 26.90 |
ENSDART00000143250
|
mlxipl
|
MLX interacting protein like |
| chr12_-_5120339 | 25.97 |
ENSDART00000168759
|
rbp4
|
retinol binding protein 4, plasma |
| chr18_-_43866001 | 25.33 |
ENSDART00000150218
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr23_+_9522942 | 25.32 |
ENSDART00000137751
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr1_-_9641845 | 25.22 |
ENSDART00000121490
ENSDART00000159411 |
ugt5b2
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B2 UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr6_-_2627488 | 24.98 |
ENSDART00000044089
ENSDART00000158333 ENSDART00000155109 |
hyi
|
hydroxypyruvate isomerase |
| chr3_-_3496738 | 24.91 |
ENSDART00000186849
|
CABZ01040998.1
|
|
| chr16_+_16969060 | 24.62 |
ENSDART00000182819
ENSDART00000191876 |
si:ch211-120k19.1
rpl18
|
si:ch211-120k19.1 ribosomal protein L18 |
| chr9_-_9977827 | 24.60 |
ENSDART00000187315
ENSDART00000010246 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr12_-_5120175 | 24.29 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr15_+_15403560 | 23.80 |
ENSDART00000049831
|
dhrs11b
|
dehydrogenase/reductase (SDR family) member 11b |
| chr20_+_2281933 | 22.85 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr22_+_21549419 | 22.74 |
ENSDART00000139411
|
plpp2b
|
phospholipid phosphatase 2b |
| chr2_-_5728843 | 22.51 |
ENSDART00000014020
|
sst2
|
somatostatin 2 |
| chr16_+_16968682 | 22.29 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr19_-_27564458 | 22.12 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr21_+_8533533 | 22.04 |
ENSDART00000077924
|
FO834888.1
|
|
| chr7_-_24520866 | 21.76 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr20_-_43741159 | 21.69 |
ENSDART00000192621
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr12_-_31457301 | 21.36 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr11_+_30296332 | 21.35 |
ENSDART00000192843
|
ugt1b7
|
UDP glucuronosyltransferase 1 family, polypeptide B7 |
| chr17_+_450956 | 21.31 |
ENSDART00000183022
ENSDART00000171386 |
zgc:194887
|
zgc:194887 |
| chr15_+_26603395 | 20.90 |
ENSDART00000188667
|
slc47a3
|
solute carrier family 47 (multidrug and toxin extrusion), member 3 |
| chr18_-_43866526 | 20.88 |
ENSDART00000111309
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr17_-_2573021 | 20.72 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr16_-_25606889 | 20.60 |
ENSDART00000077447
ENSDART00000131528 |
zgc:110410
|
zgc:110410 |
| chr10_-_11385155 | 20.33 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr8_-_8698607 | 20.12 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr15_-_1885247 | 19.57 |
ENSDART00000149703
|
porb
|
P450 (cytochrome) oxidoreductase b |
| chr24_-_26369185 | 19.53 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr17_+_15216022 | 19.46 |
ENSDART00000138831
|
gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr23_+_9522781 | 19.19 |
ENSDART00000136486
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr17_-_2578026 | 19.17 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr9_+_8380728 | 18.55 |
ENSDART00000133501
|
si:ch1073-75o15.4
|
si:ch1073-75o15.4 |
| chr16_+_54875530 | 18.32 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr21_-_30111134 | 18.27 |
ENSDART00000014223
|
slc23a1
|
solute carrier family 23 (ascorbic acid transporter), member 1 |
| chr1_-_12064715 | 18.11 |
ENSDART00000143628
ENSDART00000103406 |
pla2g12a
|
phospholipase A2, group XIIA |
| chr17_-_2595736 | 18.03 |
ENSDART00000128797
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr8_+_6576940 | 17.99 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr2_-_24407732 | 17.81 |
ENSDART00000180612
ENSDART00000153688 ENSDART00000179913 |
si:dkey-208k22.6
|
si:dkey-208k22.6 |
| chr4_-_19693978 | 17.79 |
ENSDART00000100974
ENSDART00000040405 |
snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr11_+_14281100 | 17.70 |
ENSDART00000193664
ENSDART00000161597 |
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr14_+_9009600 | 17.51 |
ENSDART00000133904
|
si:ch211-274f20.2
|
si:ch211-274f20.2 |
| chr8_+_47099033 | 16.97 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr17_-_17948587 | 16.93 |
ENSDART00000090447
|
hhipl1
|
HHIP-like 1 |
| chr17_-_2590222 | 16.88 |
ENSDART00000185711
|
CR759892.1
|
|
| chr3_-_50139860 | 16.76 |
ENSDART00000101563
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr18_-_46183462 | 16.61 |
ENSDART00000021192
|
kcnk6
|
potassium channel, subfamily K, member 6 |
| chr2_+_6067371 | 16.37 |
ENSDART00000053868
ENSDART00000145244 |
aldh9a1b
|
aldehyde dehydrogenase 9 family, member A1b |
| chr8_-_25329967 | 15.94 |
ENSDART00000139682
|
eps8l3b
|
EPS8-like 3b |
| chr6_+_53429228 | 15.24 |
ENSDART00000165067
|
abhd14b
|
abhydrolase domain containing 14B |
| chr7_-_52417060 | 15.24 |
ENSDART00000148579
|
myzap
|
myocardial zonula adherens protein |
| chr1_+_24387659 | 15.21 |
ENSDART00000130356
|
qdprb2
|
quinoid dihydropteridine reductase b2 |
| chr8_-_20838342 | 14.97 |
ENSDART00000141345
|
si:ch211-133l5.7
|
si:ch211-133l5.7 |
| chr5_-_20205075 | 14.74 |
ENSDART00000051611
|
dao.3
|
D-amino-acid oxidase, tandem duplicate 3 |
| chr22_+_29994093 | 14.10 |
ENSDART00000104778
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr8_-_33154677 | 13.85 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr12_-_33706726 | 13.69 |
ENSDART00000153135
|
myo15b
|
myosin XVB |
| chr23_+_46183410 | 13.51 |
ENSDART00000167596
ENSDART00000151149 ENSDART00000150896 |
btr31
|
bloodthirsty-related gene family, member 31 |
| chr21_+_25236297 | 13.22 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr20_-_46128590 | 12.95 |
ENSDART00000123744
|
taar1b
|
trace amine associated receptor 1b |
| chr2_-_22927958 | 12.81 |
ENSDART00000141621
|
myo7bb
|
myosin VIIBb |
| chr22_+_24645325 | 12.80 |
ENSDART00000159531
|
lpar3
|
lysophosphatidic acid receptor 3 |
| chr9_-_38036984 | 12.55 |
ENSDART00000134574
|
hacd2
|
3-hydroxyacyl-CoA dehydratase 2 |
| chr2_+_24536762 | 12.51 |
ENSDART00000144149
|
angptl4
|
angiopoietin-like 4 |
| chr22_+_15323930 | 12.34 |
ENSDART00000142416
|
si:dkey-236e20.3
|
si:dkey-236e20.3 |
| chr5_+_31959954 | 12.24 |
ENSDART00000142826
|
myo1hb
|
myosin IHb |
| chr2_-_22927581 | 12.08 |
ENSDART00000109515
|
myo7bb
|
myosin VIIBb |
| chr15_+_6459847 | 11.91 |
ENSDART00000157250
ENSDART00000065824 |
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr3_-_23643751 | 11.86 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr3_+_39099716 | 11.80 |
ENSDART00000083388
|
tmem98
|
transmembrane protein 98 |
| chr7_-_26457208 | 11.29 |
ENSDART00000173519
|
zgc:172079
|
zgc:172079 |
| chr6_-_43047774 | 11.22 |
ENSDART00000161722
|
glyctk
|
glycerate kinase |
| chr22_+_15331214 | 11.19 |
ENSDART00000136566
|
sult3st4
|
sulfotransferase family 3, cytosolic sulfotransferase 4 |
| chr4_+_1283068 | 11.01 |
ENSDART00000167233
|
chrm2a
|
cholinergic receptor, muscarinic 2a |
| chr21_-_37194669 | 10.95 |
ENSDART00000192748
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr10_+_8875195 | 10.88 |
ENSDART00000141045
|
itga2.3
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 3 |
| chr9_+_21383620 | 10.73 |
ENSDART00000130023
ENSDART00000062656 ENSDART00000180081 |
cryl1
|
crystallin, lambda 1 |
| chr20_+_5106568 | 10.65 |
ENSDART00000028039
|
cyp46a1.1
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 1 |
| chr7_+_24520518 | 10.52 |
ENSDART00000173604
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr3_+_31662126 | 10.42 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr5_+_9218318 | 10.18 |
ENSDART00000137774
|
si:ch211-12e13.1
|
si:ch211-12e13.1 |
| chr15_-_30857350 | 9.91 |
ENSDART00000138988
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr11_-_24347644 | 9.88 |
ENSDART00000089777
|
si:ch211-15p9.2
|
si:ch211-15p9.2 |
| chr19_-_18130567 | 9.50 |
ENSDART00000190659
ENSDART00000022803 |
snx10a
|
sorting nexin 10a |
| chr6_+_28051978 | 9.43 |
ENSDART00000143218
|
si:ch73-194h10.2
|
si:ch73-194h10.2 |
| chr25_-_12803723 | 9.35 |
ENSDART00000158787
|
ca5a
|
carbonic anhydrase Va |
| chr22_-_11648094 | 9.28 |
ENSDART00000191791
|
dpp4
|
dipeptidyl-peptidase 4 |
| chr5_-_44843738 | 8.82 |
ENSDART00000003926
|
fbp1a
|
fructose-1,6-bisphosphatase 1a |
| chr24_-_32150276 | 8.51 |
ENSDART00000166212
|
cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr18_-_7400075 | 8.41 |
ENSDART00000101250
|
si:dkey-30c15.13
|
si:dkey-30c15.13 |
| chr20_+_43379029 | 8.36 |
ENSDART00000142486
ENSDART00000186486 |
unc93a
|
unc-93 homolog A |
| chr12_-_20362041 | 8.25 |
ENSDART00000184145
ENSDART00000105952 |
aqp8a.2
|
aquaporin 8a, tandem duplicate 2 |
| chr21_-_13055195 | 8.12 |
ENSDART00000133517
|
myorg
|
myogenesis regulating glycosidase (putative) |
| chr18_+_26829086 | 7.96 |
ENSDART00000098356
|
slc28a1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr22_-_24992532 | 7.91 |
ENSDART00000102751
|
si:dkey-179j5.5
|
si:dkey-179j5.5 |
| chr3_-_15470944 | 7.88 |
ENSDART00000185302
|
spns1
|
spinster homolog 1 (Drosophila) |
| chr21_-_37194365 | 7.88 |
ENSDART00000100286
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr12_+_22607761 | 7.87 |
ENSDART00000153112
|
si:dkey-219e21.2
|
si:dkey-219e21.2 |
| chr25_+_5288665 | 7.80 |
ENSDART00000169540
|
CABZ01039861.1
|
|
| chr13_-_303137 | 7.79 |
ENSDART00000099131
|
chs1
|
chitin synthase 1 |
| chr20_+_39250673 | 7.62 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr11_-_30364847 | 7.58 |
ENSDART00000078387
|
pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr2_-_38261272 | 7.52 |
ENSDART00000143743
|
si:ch211-14a17.10
|
si:ch211-14a17.10 |
| chr13_+_5013572 | 6.88 |
ENSDART00000162425
|
psap
|
prosaposin |
| chr23_-_21758253 | 6.82 |
ENSDART00000046613
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr15_+_34069746 | 6.40 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr22_-_7050 | 6.39 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr7_+_26100024 | 6.36 |
ENSDART00000173726
|
si:ch211-196f2.3
|
si:ch211-196f2.3 |
| chr25_+_418932 | 6.31 |
ENSDART00000059193
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr6_+_55819038 | 5.92 |
ENSDART00000108786
|
si:ch211-81n22.1
|
si:ch211-81n22.1 |
| chr4_+_35590588 | 5.82 |
ENSDART00000183582
|
si:dkeyp-4c4.2
|
si:dkeyp-4c4.2 |
| chr6_-_35046735 | 5.81 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr18_+_26829362 | 5.80 |
ENSDART00000132728
|
slc28a1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr3_+_27770110 | 5.61 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr20_-_26937453 | 5.60 |
ENSDART00000139756
|
ftr97
|
finTRIM family, member 97 |
| chr15_+_19797918 | 5.56 |
ENSDART00000113314
|
si:ch211-229d2.5
|
si:ch211-229d2.5 |
| chr2_+_28453338 | 5.49 |
ENSDART00000020456
|
mmp15b
|
matrix metallopeptidase 15b |
| chr17_+_23926796 | 5.27 |
ENSDART00000021177
|
pex13
|
peroxisomal biogenesis factor 13 |
| chr5_+_32815745 | 5.11 |
ENSDART00000181535
|
crata
|
carnitine O-acetyltransferase a |
| chr7_+_73444325 | 5.02 |
ENSDART00000123016
|
si:ch211-142d6.2
|
si:ch211-142d6.2 |
| chr10_+_8197827 | 4.90 |
ENSDART00000026244
|
mtrex
|
Mtr4 exosome RNA helicase |
| chr19_-_27564980 | 4.82 |
ENSDART00000171967
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr5_-_24112175 | 4.77 |
ENSDART00000131595
|
si:ch211-114c12.5
|
si:ch211-114c12.5 |
| chr2_+_47249179 | 4.65 |
ENSDART00000144438
|
si:ch211-284d12.3
|
si:ch211-284d12.3 |
| chr22_-_16400484 | 4.60 |
ENSDART00000135987
|
lama3
|
laminin, alpha 3 |
| chr20_-_26936887 | 4.46 |
ENSDART00000160827
|
ftr79
|
finTRIM family, member 79 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hnf4a+hnf4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 36.5 | 109.5 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 22.6 | 67.9 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 15.4 | 46.2 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 13.9 | 139.5 | GO:1990402 | embryonic liver development(GO:1990402) |
| 13.4 | 66.8 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 11.0 | 88.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 9.4 | 37.5 | GO:0030091 | protein repair(GO:0030091) |
| 7.5 | 59.7 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 7.2 | 50.3 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 7.1 | 21.4 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 6.9 | 27.4 | GO:0046440 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 6.3 | 18.8 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 6.1 | 18.3 | GO:0015882 | L-ascorbic acid transport(GO:0015882) |
| 5.1 | 86.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 4.3 | 12.8 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 4.2 | 58.9 | GO:0016556 | mRNA modification(GO:0016556) |
| 3.7 | 14.7 | GO:0046144 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 3.6 | 317.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 3.6 | 25.0 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 3.5 | 55.3 | GO:0071715 | icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) |
| 3.2 | 116.3 | GO:0050821 | protein stabilization(GO:0050821) |
| 3.1 | 40.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 3.0 | 72.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 2.8 | 11.0 | GO:0010459 | cardiac conduction system development(GO:0003161) negative regulation of heart rate(GO:0010459) |
| 2.7 | 15.9 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 2.5 | 7.6 | GO:1903173 | fatty alcohol metabolic process(GO:1903173) |
| 2.5 | 9.9 | GO:1904182 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 2.3 | 16.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 2.2 | 32.4 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 2.1 | 51.5 | GO:0032355 | response to estradiol(GO:0032355) |
| 2.0 | 86.2 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 1.9 | 25.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 1.9 | 9.5 | GO:0070986 | left/right axis specification(GO:0070986) |
| 1.7 | 48.5 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 1.6 | 12.5 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 1.6 | 65.6 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 1.6 | 7.8 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 1.5 | 10.6 | GO:0006707 | steroid catabolic process(GO:0006706) cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 1.5 | 15.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 1.4 | 218.0 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 1.2 | 11.9 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 1.1 | 3.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 1.0 | 5.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 1.0 | 11.9 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 1.0 | 6.8 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.9 | 49.4 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.9 | 13.8 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.9 | 18.3 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.9 | 42.3 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.9 | 7.9 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.8 | 3.4 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.8 | 112.2 | GO:0055123 | digestive system development(GO:0055123) |
| 0.7 | 50.4 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.6 | 61.6 | GO:0006956 | complement activation(GO:0006956) protein activation cascade(GO:0072376) |
| 0.6 | 9.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.6 | 4.2 | GO:0032475 | otolith formation(GO:0032475) |
| 0.6 | 2.9 | GO:2000562 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.5 | 5.3 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.5 | 48.1 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.5 | 12.6 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.4 | 12.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.4 | 10.6 | GO:0051923 | sulfation(GO:0051923) |
| 0.4 | 2.9 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.3 | 3.7 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.3 | 4.2 | GO:0030534 | adult behavior(GO:0030534) |
| 0.3 | 11.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.3 | 22.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.2 | 49.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.2 | 2.8 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.2 | 1.0 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.2 | 5.5 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.2 | 4.9 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 3.2 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 28.0 | GO:0006869 | lipid transport(GO:0006869) |
| 0.1 | 4.1 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 16.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 1.0 | GO:0042664 | negative regulation of cell fate specification(GO:0009996) negative regulation of endodermal cell fate specification(GO:0042664) |
| 0.1 | 6.9 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 61.1 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.1 | 3.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 2.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 4.0 | GO:0097191 | extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.1 | 11.5 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.1 | 19.6 | GO:0009725 | response to hormone(GO:0009725) |
| 0.1 | 0.6 | GO:0098927 | protein targeting to lysosome(GO:0006622) early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 0.3 | GO:0006549 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.1 | 4.6 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 1.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 7.6 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 3.7 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 7.1 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 2.5 | GO:0006401 | RNA catabolic process(GO:0006401) |
| 0.0 | 6.4 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 2.8 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 2.5 | GO:0007187 | G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger(GO:0007187) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.2 | 53.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 4.5 | 67.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 2.2 | 17.8 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 1.8 | 20.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 1.8 | 5.3 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 1.5 | 9.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 1.2 | 3.7 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 1.0 | 7.8 | GO:0030428 | cell septum(GO:0030428) |
| 0.7 | 24.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.7 | 889.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.5 | 3.3 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.5 | 55.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.5 | 328.9 | GO:0005576 | extracellular region(GO:0005576) |
| 0.5 | 20.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.4 | 15.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.3 | 19.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.3 | 4.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.3 | 24.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 109.2 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.2 | 163.3 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.2 | 103.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 5.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 4.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 819.8 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.1 | 0.6 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 45.4 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 2.9 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 29.1 | 116.3 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 22.0 | 88.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 15.4 | 46.2 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 14.3 | 42.8 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 9.4 | 37.5 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 8.0 | 63.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 7.8 | 86.0 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 7.4 | 51.5 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 7.2 | 50.3 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 6.8 | 40.7 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 5.6 | 45.2 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 5.5 | 16.4 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 4.9 | 19.6 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 4.6 | 32.4 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 4.6 | 27.4 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 4.2 | 245.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 4.0 | 71.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 3.8 | 15.2 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 3.7 | 14.7 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 3.1 | 12.6 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 2.9 | 8.8 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 2.8 | 67.9 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 2.8 | 42.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 2.8 | 11.2 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 2.6 | 297.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 2.5 | 9.9 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 2.4 | 59.7 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 1.9 | 21.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 1.8 | 10.6 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 1.6 | 63.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 1.6 | 54.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 1.5 | 30.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 1.5 | 40.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 1.4 | 18.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 1.4 | 40.4 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 1.4 | 5.6 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 1.3 | 18.4 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 1.3 | 5.1 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 1.2 | 77.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 1.2 | 8.2 | GO:0015250 | water channel activity(GO:0015250) |
| 1.2 | 5.8 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 1.1 | 37.8 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 1.1 | 3.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 1.0 | 88.6 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 1.0 | 11.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 1.0 | 7.8 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.9 | 22.7 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.9 | 109.5 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.9 | 12.8 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.8 | 4.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.8 | 20.7 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.8 | 10.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.8 | 53.4 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.6 | 11.9 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.5 | 9.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.5 | 9.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.5 | 3.4 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.4 | 101.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.4 | 21.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.4 | 62.1 | GO:0016798 | hydrolase activity, acting on glycosyl bonds(GO:0016798) |
| 0.4 | 3.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.4 | 16.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.4 | 2.8 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.3 | 19.5 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.3 | 52.2 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.3 | 197.1 | GO:0008289 | lipid binding(GO:0008289) |
| 0.3 | 2.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.3 | 7.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.3 | 53.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.2 | 13.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.2 | 3.7 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.2 | 104.9 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.2 | 13.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 14.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 8.9 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 22.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 3.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 24.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 11.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 3.9 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.1 | 4.0 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.1 | 57.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 2.7 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 6.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 8.1 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.1 | 4.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 1.2 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 28.5 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 5.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 4.4 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 6.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 24.3 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 10.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.5 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 112.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 1.8 | 116.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.5 | 3.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.4 | 64.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.4 | 4.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.3 | 49.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.3 | 17.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.3 | 11.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.3 | 6.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 12.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 3.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 12.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.6 | 180.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 8.4 | 66.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 7.7 | 46.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 7.2 | 57.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 6.4 | 76.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 4.5 | 18.1 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 4.3 | 34.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 4.3 | 29.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 2.8 | 16.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 2.7 | 48.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 2.7 | 21.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 2.1 | 18.8 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 1.1 | 19.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 1.1 | 71.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.8 | 9.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.7 | 12.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.6 | 52.9 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.4 | 5.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.4 | 18.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.3 | 3.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 49.6 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.2 | 24.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 4.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 6.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 12.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 7.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 10.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 6.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |