Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
Results for grhl1
Z-value: 4.15
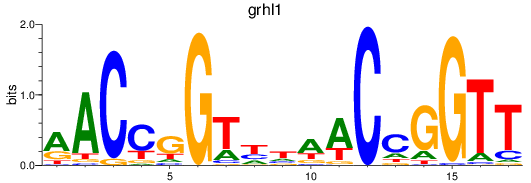
Transcription factors associated with grhl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
grhl1
|
ENSDARG00000061391 | grainyhead-like transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| grhl1 | dr11_v1_chr17_-_32413147_32413147 | 0.86 | 4.0e-28 | Click! |
Activity profile of grhl1 motif
Sorted Z-values of grhl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_12360409 | 87.74 |
ENSDART00000164596
|
tmprss13a
|
transmembrane protease, serine 13a |
| chr9_-_30264415 | 86.84 |
ENSDART00000060150
|
mid1ip1a
|
MID1 interacting protein 1a |
| chr15_+_42560354 | 79.94 |
ENSDART00000059484
|
CLDN8 (1 of many)
|
zgc:110333 |
| chr16_-_9675982 | 75.55 |
ENSDART00000113724
|
mal2
|
mal, T cell differentiation protein 2 (gene/pseudogene) |
| chr12_-_28794957 | 74.02 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr21_+_11244068 | 71.31 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr7_+_22657566 | 69.46 |
ENSDART00000141048
|
ponzr5
|
plac8 onzin related protein 5 |
| chr24_+_38671054 | 65.02 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr8_-_24252933 | 59.77 |
ENSDART00000057624
|
zgc:110353
|
zgc:110353 |
| chr10_-_36793412 | 59.37 |
ENSDART00000185966
|
dhrs13a.2
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 2 |
| chr6_+_23057311 | 57.50 |
ENSDART00000026448
|
evpla
|
envoplakin a |
| chr16_+_35905031 | 56.36 |
ENSDART00000162411
|
sh3d21
|
SH3 domain containing 21 |
| chr22_+_19552987 | 56.13 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr8_+_3405612 | 55.50 |
ENSDART00000163437
|
zgc:112433
|
zgc:112433 |
| chr17_+_28102487 | 54.69 |
ENSDART00000131638
ENSDART00000076265 |
zgc:91908
|
zgc:91908 |
| chr15_+_41919484 | 54.27 |
ENSDART00000099821
ENSDART00000146246 |
nlrp16
|
NACHT, LRR and PYD domains-containing protein 16 |
| chr5_-_57686487 | 49.39 |
ENSDART00000074264
|
crfb12
|
cytokine receptor family member B12 |
| chr3_-_16537441 | 49.13 |
ENSDART00000080749
ENSDART00000133824 |
eps8l1
|
eps8-like1 |
| chr5_-_37900350 | 47.86 |
ENSDART00000084839
ENSDART00000084841 ENSDART00000133437 |
tmprss13b
|
transmembrane protease, serine 13b |
| chr16_+_17715243 | 47.65 |
ENSDART00000149437
ENSDART00000149596 |
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr23_-_22650668 | 45.42 |
ENSDART00000132733
|
ca6
|
carbonic anhydrase VI |
| chr5_-_6567464 | 45.25 |
ENSDART00000184985
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr4_-_9780931 | 44.75 |
ENSDART00000134280
ENSDART00000150664 ENSDART00000150304 ENSDART00000080744 |
svopl
|
SVOP-like |
| chr22_+_25672155 | 44.23 |
ENSDART00000087769
|
si:ch211-250e5.2
|
si:ch211-250e5.2 |
| chr10_-_22127942 | 44.19 |
ENSDART00000133374
|
ponzr2
|
plac8 onzin related protein 2 |
| chr15_+_22014029 | 43.98 |
ENSDART00000079504
|
ankk1
|
ankyrin repeat and kinase domain containing 1 |
| chr23_-_22650992 | 42.44 |
ENSDART00000079007
|
ca6
|
carbonic anhydrase VI |
| chr22_+_22021936 | 41.93 |
ENSDART00000149586
|
gna15.1
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 1 |
| chr16_-_31976269 | 41.04 |
ENSDART00000139664
|
styk1
|
serine/threonine/tyrosine kinase 1 |
| chr5_+_58665648 | 40.73 |
ENSDART00000167481
|
zgc:194948
|
zgc:194948 |
| chr19_-_32500373 | 40.72 |
ENSDART00000052104
|
fuca1.1
|
alpha-L-fucosidase 1, tandem duplicate 1 |
| chr13_+_42544009 | 40.58 |
ENSDART00000145409
|
si:dkey-221j11.3
|
si:dkey-221j11.3 |
| chr16_+_23431189 | 40.58 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr3_-_18756076 | 39.83 |
ENSDART00000055766
|
zgc:113333
|
zgc:113333 |
| chr7_+_19835569 | 39.71 |
ENSDART00000149812
|
ovol1a
|
ovo-like zinc finger 1a |
| chr4_+_76722754 | 39.48 |
ENSDART00000153867
|
ms4a17a.3
|
membrane-spanning 4-domains, subfamily A, member 17A.3 |
| chr1_-_48933 | 38.08 |
ENSDART00000171162
|
ildr1a
|
immunoglobulin-like domain containing receptor 1a |
| chr3_-_18755651 | 37.59 |
ENSDART00000145277
|
zgc:113333
|
zgc:113333 |
| chr8_-_36399884 | 36.87 |
ENSDART00000108538
|
si:zfos-2070c2.3
|
si:zfos-2070c2.3 |
| chr2_+_5521671 | 36.75 |
ENSDART00000099647
ENSDART00000138443 |
crfb16
|
cytokine receptor family member B16 |
| chr19_-_41472228 | 35.27 |
ENSDART00000113388
|
dlx5a
|
distal-less homeobox 5a |
| chr3_-_29977495 | 31.40 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr21_+_25625026 | 31.29 |
ENSDART00000134678
|
ovol1b
|
ovo-like zinc finger 1b |
| chr2_-_47620806 | 30.41 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr21_-_3700334 | 28.98 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr25_-_24202576 | 28.78 |
ENSDART00000048507
|
uevld
|
UEV and lactate/malate dehyrogenase domains |
| chr22_-_22337382 | 28.46 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr21_-_13668358 | 27.78 |
ENSDART00000180323
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr5_-_29570141 | 27.75 |
ENSDART00000043259
|
entpd2a.2
|
ectonucleoside triphosphate diphosphohydrolase 2a, tandem duplicate 2 |
| chr4_-_26413391 | 26.76 |
ENSDART00000145955
|
b4galnt3a
|
beta-1,4-N-acetyl-galactosaminyl transferase 3a |
| chr6_+_30668098 | 26.40 |
ENSDART00000112294
|
ttc22
|
tetratricopeptide repeat domain 22 |
| chr10_+_4924388 | 26.25 |
ENSDART00000108595
|
slc46a2
|
solute carrier family 46 member 2 |
| chr13_+_18533005 | 24.31 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr12_+_30360579 | 22.89 |
ENSDART00000152900
|
si:ch211-225b10.3
|
si:ch211-225b10.3 |
| chr16_+_7154753 | 22.67 |
ENSDART00000163281
ENSDART00000168274 |
bmper
|
BMP binding endothelial regulator |
| chr16_-_28091597 | 22.58 |
ENSDART00000166595
|
CR626941.1
|
|
| chr17_+_22102791 | 22.20 |
ENSDART00000047772
|
mal
|
mal, T cell differentiation protein |
| chr14_+_146857 | 22.11 |
ENSDART00000122521
|
CABZ01088229.1
|
|
| chr4_-_12725513 | 21.37 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr16_+_53526135 | 21.24 |
ENSDART00000083558
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr7_-_24193051 | 21.20 |
ENSDART00000169737
ENSDART00000171085 |
si:ch211-216p19.6
|
si:ch211-216p19.6 |
| chr15_+_509126 | 20.10 |
ENSDART00000102274
|
ftr86
|
finTRIM family, member 86 |
| chr4_+_4232562 | 19.85 |
ENSDART00000177529
|
smkr1
|
small lysine rich protein 1 |
| chr2_-_58075414 | 19.80 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr9_-_40014339 | 19.63 |
ENSDART00000166918
|
IKZF2
|
si:zfos-1425h8.1 |
| chr21_+_5801105 | 16.53 |
ENSDART00000151225
ENSDART00000184487 |
ccng2
|
cyclin G2 |
| chr12_+_31673588 | 13.76 |
ENSDART00000152971
|
dnmbp
|
dynamin binding protein |
| chr15_+_41901970 | 13.73 |
ENSDART00000152724
|
si:ch211-191a16.5
|
si:ch211-191a16.5 |
| chr9_-_14108896 | 13.65 |
ENSDART00000135209
|
prkag3b
|
protein kinase, AMP-activated, gamma 3b non-catalytic subunit |
| chr7_-_56606752 | 13.20 |
ENSDART00000138714
|
sult5a1
|
sulfotransferase family 5A, member 1 |
| chr3_+_34140507 | 12.52 |
ENSDART00000131802
|
si:dkey-204f11.64
|
si:dkey-204f11.64 |
| chr14_+_45702521 | 11.36 |
ENSDART00000112470
|
ccdc88b
|
coiled-coil domain containing 88B |
| chr8_-_13029297 | 10.86 |
ENSDART00000144305
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr2_-_17827983 | 10.05 |
ENSDART00000166518
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr15_+_41901710 | 10.04 |
ENSDART00000159874
|
si:ch211-191a16.5
|
si:ch211-191a16.5 |
| chr25_+_15994100 | 9.83 |
ENSDART00000144723
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr13_-_30161684 | 9.42 |
ENSDART00000040409
|
ppa1b
|
pyrophosphatase (inorganic) 1b |
| chr19_-_10432134 | 8.72 |
ENSDART00000081440
|
il11b
|
interleukin 11b |
| chr3_-_32831429 | 8.63 |
ENSDART00000184932
|
zgc:153733
|
zgc:153733 |
| chr4_-_14954327 | 8.54 |
ENSDART00000182729
|
slc26a5
|
solute carrier family 26 (anion exchanger), member 5 |
| chr3_+_37196258 | 8.33 |
ENSDART00000187944
ENSDART00000185896 |
fmnl1a
|
formin-like 1a |
| chr12_+_4036409 | 8.18 |
ENSDART00000106650
|
zgc:123217
|
zgc:123217 |
| chr12_+_30705769 | 7.12 |
ENSDART00000186448
ENSDART00000066259 |
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr3_-_32831971 | 7.06 |
ENSDART00000075270
|
zgc:153733
|
zgc:153733 |
| chr12_+_30706158 | 6.99 |
ENSDART00000133869
|
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr1_-_31105376 | 6.65 |
ENSDART00000132466
|
ppp1r9alb
|
protein phosphatase 1 regulatory subunit 9A-like B |
| chr14_-_33945692 | 6.53 |
ENSDART00000168546
ENSDART00000189778 |
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr22_-_14361022 | 6.40 |
ENSDART00000140224
|
si:dkeyp-122a9.2
|
si:dkeyp-122a9.2 |
| chr4_-_14954029 | 6.10 |
ENSDART00000038642
|
slc26a5
|
solute carrier family 26 (anion exchanger), member 5 |
| chr1_-_47161996 | 5.12 |
ENSDART00000053153
|
mhc1zba
|
major histocompatibility complex class I ZBA |
| chr16_-_10837245 | 5.09 |
ENSDART00000036891
|
rabac1
|
Rab acceptor 1 (prenylated) |
| chr15_+_29276508 | 4.64 |
ENSDART00000170537
ENSDART00000126559 |
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr15_-_5901514 | 4.63 |
ENSDART00000155252
|
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr23_-_19225709 | 3.88 |
ENSDART00000080099
|
oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr2_-_58201173 | 3.79 |
ENSDART00000166282
|
pnp5b
|
purine nucleoside phosphorylase 5b |
| chr24_+_42074143 | 3.73 |
ENSDART00000170514
|
top1mt
|
DNA topoisomerase I mitochondrial |
| chr11_-_42417194 | 3.69 |
ENSDART00000191086
ENSDART00000076650 ENSDART00000165903 ENSDART00000104444 |
slmapa
|
sarcolemma associated protein a |
| chr11_-_45140227 | 3.56 |
ENSDART00000184062
|
cant1b
|
calcium activated nucleotidase 1b |
| chr11_-_17964525 | 3.38 |
ENSDART00000018948
|
cishb
|
cytokine inducible SH2-containing protein b |
| chr1_-_9858508 | 2.28 |
ENSDART00000147904
|
mad1l1
|
mitotic arrest deficient 1 like 1 |
| chr5_+_4006837 | 2.24 |
ENSDART00000138862
|
pigw
|
phosphatidylinositol glycan anchor biosynthesis, class W |
| chr24_-_8412526 | 1.67 |
ENSDART00000150055
|
slc35b3
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B3 |
| chr3_-_30186296 | 0.44 |
ENSDART00000134395
ENSDART00000077057 ENSDART00000017422 |
tbc1d17
|
TBC1 domain family, member 17 |
| chr20_+_20731052 | 0.33 |
ENSDART00000047662
|
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr8_+_8459192 | 0.29 |
ENSDART00000140942
ENSDART00000014939 |
comta
|
catechol-O-methyltransferase a |
| chr22_-_13649588 | 0.28 |
ENSDART00000131877
|
si:ch211-279g13.1
|
si:ch211-279g13.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of grhl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.1 | 75.5 | GO:0045056 | transcytosis(GO:0045056) |
| 6.8 | 40.7 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 5.7 | 28.5 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 5.7 | 39.7 | GO:0070285 | pigment cell development(GO:0070285) |
| 4.6 | 27.7 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 4.4 | 35.3 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 3.7 | 14.6 | GO:0019532 | oxalate transport(GO:0019532) |
| 3.1 | 59.4 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 2.1 | 21.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 1.9 | 41.9 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 1.7 | 87.9 | GO:0036269 | swimming behavior(GO:0036269) |
| 1.5 | 86.8 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 1.3 | 15.7 | GO:0001881 | receptor recycling(GO:0001881) |
| 1.3 | 57.5 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 1.1 | 10.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.9 | 29.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.6 | 21.9 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.6 | 49.4 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.6 | 14.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.5 | 22.9 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.5 | 13.2 | GO:0051923 | sulfation(GO:0051923) |
| 0.4 | 13.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.3 | 16.5 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.3 | 31.3 | GO:0009913 | epidermal cell differentiation(GO:0009913) |
| 0.3 | 66.5 | GO:0006869 | lipid transport(GO:0006869) |
| 0.3 | 2.3 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.2 | 3.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.2 | 11.4 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.2 | 6.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 9.8 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 95.4 | GO:0005975 | carbohydrate metabolic process(GO:0005975) |
| 0.1 | 8.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 24.2 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.1 | 143.8 | GO:0006508 | proteolysis(GO:0006508) |
| 0.1 | 2.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 4.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 6.6 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 68.2 | GO:0015031 | protein transport(GO:0015031) |
| 0.1 | 13.8 | GO:0072001 | kidney development(GO:0001822) renal system development(GO:0072001) |
| 0.0 | 3.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.3 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 54.5 | GO:0035556 | intracellular signal transduction(GO:0035556) |
| 0.0 | 3.7 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 23.7 | GO:0006468 | protein phosphorylation(GO:0006468) |
| 0.0 | 56.9 | GO:0006357 | regulation of transcription from RNA polymerase II promoter(GO:0006357) |
| 0.0 | 5.1 | GO:0006955 | immune response(GO:0006955) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 74.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 1.1 | 13.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.9 | 22.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.8 | 54.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.7 | 75.5 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.5 | 57.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.5 | 21.2 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.5 | 26.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.4 | 16.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.3 | 14.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.3 | 58.4 | GO:0098791 | Golgi subcompartment(GO:0098791) |
| 0.3 | 30.4 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.2 | 93.0 | GO:0009986 | cell surface(GO:0009986) |
| 0.2 | 59.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.2 | 59.4 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 38.5 | GO:0005764 | lysosome(GO:0005764) |
| 0.1 | 9.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 6.6 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 115.7 | GO:0005576 | extracellular region(GO:0005576) |
| 0.1 | 69.9 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.1 | 3.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 11.4 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 369.5 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 2.3 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 171.3 | GO:0005634 | nucleus(GO:0005634) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.6 | 40.7 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 9.6 | 57.5 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 8.6 | 77.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 5.9 | 59.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 5.4 | 26.8 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 5.0 | 75.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 4.7 | 41.9 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 3.1 | 87.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 2.7 | 74.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 2.4 | 9.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 2.2 | 56.1 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 1.6 | 27.7 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 1.5 | 21.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 1.4 | 40.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.9 | 14.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.9 | 3.7 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.8 | 14.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.7 | 47.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.7 | 86.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.6 | 21.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.6 | 12.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.6 | 22.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.6 | 1.7 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.5 | 13.7 | GO:0016208 | AMP binding(GO:0016208) |
| 0.5 | 28.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.5 | 95.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.4 | 3.8 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.4 | 3.6 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.3 | 11.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.3 | 16.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 28.8 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.2 | 4.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 29.3 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.2 | 13.2 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 6.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 3.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 10.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 65.6 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.1 | 24.6 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 165.8 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 8.7 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 15.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.2 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 31.4 | GO:0016491 | oxidoreductase activity(GO:0016491) |
| 0.0 | 39.3 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.0 | 40.7 | GO:0022857 | transmembrane transporter activity(GO:0022857) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 5.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 13.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 22.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 87.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 2.8 | 45.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.6 | 29.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 2.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |