Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
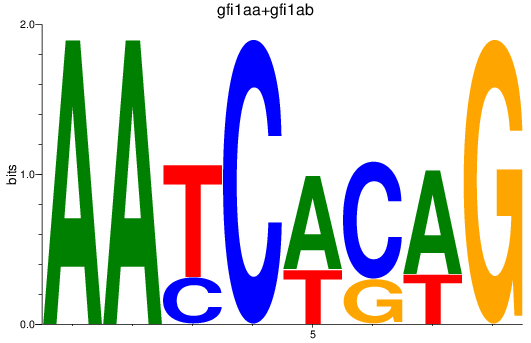
Results for gfi1aa+gfi1ab
Z-value: 2.37
Transcription factors associated with gfi1aa+gfi1ab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gfi1aa
|
ENSDARG00000020746 | growth factor independent 1A transcription repressor a |
|
gfi1ab
|
ENSDARG00000044457 | growth factor independent 1A transcription repressor b |
|
gfi1ab
|
ENSDARG00000114140 | growth factor independent 1A transcription repressor b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gfi1aa | dr11_v1_chr2_+_10771787_10771787 | 0.77 | 1.1e-19 | Click! |
| gfi1ab | dr11_v1_chr6_-_29007493_29007493 | -0.09 | 4.0e-01 | Click! |
Activity profile of gfi1aa+gfi1ab motif
Sorted Z-values of gfi1aa+gfi1ab motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_15433671 | 43.08 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr17_+_15433518 | 42.74 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr9_-_18877597 | 34.13 |
ENSDART00000099446
|
kctd4
|
potassium channel tetramerization domain containing 4 |
| chr18_-_1185772 | 33.40 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr18_+_22793465 | 30.77 |
ENSDART00000149685
|
gnao1a
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a |
| chr21_+_26697536 | 30.66 |
ENSDART00000004109
|
gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr14_+_35806605 | 30.29 |
ENSDART00000173093
|
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr5_+_64732270 | 24.44 |
ENSDART00000134241
|
olfm1a
|
olfactomedin 1a |
| chr15_-_24869826 | 23.88 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr7_-_22132265 | 22.45 |
ENSDART00000125284
ENSDART00000112978 |
nlgn2a
|
neuroligin 2a |
| chr6_+_40354424 | 20.55 |
ENSDART00000047416
|
slc4a8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr24_-_8729531 | 20.44 |
ENSDART00000082346
|
tfap2a
|
transcription factor AP-2 alpha |
| chr16_-_29437373 | 19.81 |
ENSDART00000148405
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr3_+_40170216 | 18.73 |
ENSDART00000011568
|
syngr3a
|
synaptogyrin 3a |
| chr3_-_36440705 | 18.64 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr17_-_36936649 | 18.54 |
ENSDART00000145236
|
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr25_+_21829777 | 18.40 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr1_-_31505144 | 18.01 |
ENSDART00000087115
|
rims1b
|
regulating synaptic membrane exocytosis 1b |
| chr2_-_16562505 | 17.65 |
ENSDART00000156406
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr20_+_18580176 | 17.52 |
ENSDART00000185310
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr8_-_14052349 | 17.21 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr3_+_54168007 | 17.20 |
ENSDART00000109894
|
olfm2a
|
olfactomedin 2a |
| chr4_+_8168514 | 16.93 |
ENSDART00000150830
|
ninj2
|
ninjurin 2 |
| chr4_+_11375894 | 16.76 |
ENSDART00000190471
ENSDART00000143963 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr17_-_36936856 | 16.70 |
ENSDART00000010274
ENSDART00000188887 |
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr17_+_26569601 | 16.45 |
ENSDART00000153897
|
ndnfl
|
neuron-derived neurotrophic factor , like |
| chr18_+_26895994 | 16.34 |
ENSDART00000098347
|
ch25hl1.2
|
cholesterol 25-hydroxylase like 1, tandem duplicate 2 |
| chr18_-_38088099 | 16.00 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr24_-_29997145 | 15.86 |
ENSDART00000135094
|
palmdb
|
palmdelphin b |
| chr17_+_12698532 | 15.78 |
ENSDART00000064509
ENSDART00000136830 |
stmn4l
|
stathmin-like 4, like |
| chr17_-_12385308 | 15.77 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr7_-_51546386 | 15.68 |
ENSDART00000174306
|
nhsl2
|
NHS-like 2 |
| chr15_-_19250543 | 15.39 |
ENSDART00000092705
ENSDART00000138895 |
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr13_+_27951688 | 15.35 |
ENSDART00000050303
|
b3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr22_+_18389271 | 15.03 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr15_+_36115955 | 14.98 |
ENSDART00000032702
|
sst1.2
|
somatostatin 1, tandem duplicate 2 |
| chr13_+_51579851 | 14.76 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr17_-_8638713 | 14.75 |
ENSDART00000148971
|
ctbp2a
|
C-terminal binding protein 2a |
| chr2_-_31735142 | 14.34 |
ENSDART00000130903
|
ralyl
|
RALY RNA binding protein like |
| chr22_-_11493236 | 14.33 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr7_-_26408472 | 14.24 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr13_-_27660955 | 14.19 |
ENSDART00000188651
ENSDART00000134494 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr4_+_6643421 | 14.12 |
ENSDART00000099462
|
gpr85
|
G protein-coupled receptor 85 |
| chr15_+_22311803 | 14.12 |
ENSDART00000150182
|
hepacama
|
hepatic and glial cell adhesion molecule a |
| chr10_+_23022263 | 13.98 |
ENSDART00000138955
|
si:dkey-175g6.2
|
si:dkey-175g6.2 |
| chr14_-_40389699 | 13.96 |
ENSDART00000181581
ENSDART00000173398 |
pcdh19
|
protocadherin 19 |
| chr23_-_29505463 | 13.79 |
ENSDART00000050915
|
kif1b
|
kinesin family member 1B |
| chr16_+_39159752 | 13.45 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr13_+_15816573 | 13.39 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr23_-_21453614 | 13.31 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr4_-_7212875 | 12.90 |
ENSDART00000161297
|
lrrn3b
|
leucine rich repeat neuronal 3b |
| chr7_+_26629084 | 12.90 |
ENSDART00000101044
ENSDART00000173765 |
hsbp1a
|
heat shock factor binding protein 1a |
| chr5_+_58372164 | 12.89 |
ENSDART00000057910
|
nrgna
|
neurogranin (protein kinase C substrate, RC3) a |
| chr19_-_30562648 | 12.84 |
ENSDART00000171006
|
hpcal4
|
hippocalcin like 4 |
| chr23_+_19594608 | 12.83 |
ENSDART00000134865
|
slmapb
|
sarcolemma associated protein b |
| chr19_-_15335787 | 12.65 |
ENSDART00000187131
|
hivep3a
|
human immunodeficiency virus type I enhancer binding protein 3a |
| chr7_+_38716048 | 12.65 |
ENSDART00000024590
|
syt13
|
synaptotagmin XIII |
| chr5_-_35252761 | 12.58 |
ENSDART00000051278
|
tnpo1
|
transportin 1 |
| chr12_+_25600685 | 12.55 |
ENSDART00000077157
|
six3b
|
SIX homeobox 3b |
| chr9_+_23255410 | 12.48 |
ENSDART00000113241
ENSDART00000137231 |
tmem163a
|
transmembrane protein 163a |
| chr23_+_41799748 | 12.47 |
ENSDART00000144257
|
pdyn
|
prodynorphin |
| chr15_+_5901970 | 12.46 |
ENSDART00000114134
|
wrb
|
tryptophan rich basic protein |
| chr1_+_14283692 | 12.41 |
ENSDART00000017679
|
ppp2r2ca
|
protein phosphatase 2, regulatory subunit B, gamma a |
| chr17_-_17130942 | 12.41 |
ENSDART00000064241
|
nrxn3a
|
neurexin 3a |
| chr2_-_9915814 | 12.32 |
ENSDART00000091644
ENSDART00000177556 |
abi1b
|
abl-interactor 1b |
| chr15_-_44601331 | 12.29 |
ENSDART00000161514
|
zgc:165508
|
zgc:165508 |
| chr7_-_27686021 | 12.26 |
ENSDART00000079112
ENSDART00000100989 |
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr6_+_40523370 | 12.17 |
ENSDART00000033819
|
prkcda
|
protein kinase C, delta a |
| chr25_-_19090479 | 12.13 |
ENSDART00000027465
ENSDART00000177670 |
cacna2d4b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4b |
| chr10_-_34871737 | 11.95 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr25_+_19999623 | 11.90 |
ENSDART00000026401
|
zgc:194665
|
zgc:194665 |
| chr24_+_14713776 | 11.81 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr11_-_4235811 | 11.74 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr4_+_17336557 | 11.74 |
ENSDART00000111650
|
pmch
|
pro-melanin-concentrating hormone |
| chr10_+_29431529 | 11.71 |
ENSDART00000158154
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr5_-_10946232 | 11.68 |
ENSDART00000163139
ENSDART00000031265 |
rtn4r
|
reticulon 4 receptor |
| chr23_+_22200467 | 11.58 |
ENSDART00000025414
|
slc2a1a
|
solute carrier family 2 (facilitated glucose transporter), member 1a |
| chr6_-_14040136 | 11.51 |
ENSDART00000065361
ENSDART00000179765 |
etv5b
|
ets variant 5b |
| chr25_-_15049694 | 11.44 |
ENSDART00000162485
ENSDART00000164384 ENSDART00000165632 ENSDART00000159490 |
pax6a
|
paired box 6a |
| chr22_-_20575679 | 11.36 |
ENSDART00000089033
|
lingo3a
|
leucine rich repeat and Ig domain containing 3a |
| chr5_-_23362602 | 11.34 |
ENSDART00000137120
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr14_-_32403554 | 11.33 |
ENSDART00000172873
ENSDART00000173408 ENSDART00000173114 ENSDART00000185594 ENSDART00000186762 ENSDART00000010982 |
fgf13a
|
fibroblast growth factor 13a |
| chr13_-_27767330 | 11.30 |
ENSDART00000131631
ENSDART00000112553 ENSDART00000189911 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr10_-_27046639 | 11.28 |
ENSDART00000041841
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr12_+_21299338 | 11.24 |
ENSDART00000074540
ENSDART00000133188 |
ca10a
|
carbonic anhydrase Xa |
| chr2_+_22702488 | 11.10 |
ENSDART00000076647
|
kif1ab
|
kinesin family member 1Ab |
| chr24_-_6501211 | 11.08 |
ENSDART00000186241
ENSDART00000109040 ENSDART00000136154 |
gpr158a
|
G protein-coupled receptor 158a |
| chr14_+_49135264 | 11.04 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr5_+_70155935 | 11.01 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr21_-_4032650 | 10.90 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr3_-_35800221 | 10.82 |
ENSDART00000031390
|
caskin1
|
CASK interacting protein 1 |
| chr7_-_23996133 | 10.80 |
ENSDART00000173761
|
si:dkey-183c6.8
|
si:dkey-183c6.8 |
| chr6_-_42073367 | 10.79 |
ENSDART00000154304
|
grm2a
|
glutamate receptor, metabotropic 2a |
| chr10_-_22057001 | 10.67 |
ENSDART00000016575
|
tlx3b
|
T cell leukemia homeobox 3b |
| chr14_-_40390757 | 10.52 |
ENSDART00000149443
|
pcdh19
|
protocadherin 19 |
| chr10_+_21722892 | 10.49 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr5_+_43965078 | 10.29 |
ENSDART00000113502
ENSDART00000187143 |
TMEM8B
|
si:dkey-84j12.1 |
| chr11_-_23080970 | 10.25 |
ENSDART00000127791
|
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr9_-_27442339 | 10.11 |
ENSDART00000138602
|
stxbp5l
|
syntaxin binding protein 5-like |
| chr17_-_26911852 | 10.08 |
ENSDART00000045842
|
rcan3
|
regulator of calcineurin 3 |
| chr2_-_3678029 | 9.94 |
ENSDART00000146861
|
mmp16b
|
matrix metallopeptidase 16b (membrane-inserted) |
| chr19_-_9503473 | 9.88 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr25_-_7520937 | 9.87 |
ENSDART00000170050
|
cdkn1cb
|
cyclin-dependent kinase inhibitor 1Cb |
| chr25_-_13381854 | 9.86 |
ENSDART00000164621
ENSDART00000169129 |
ndrg4
|
NDRG family member 4 |
| chr20_-_29864390 | 9.79 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr12_+_19305390 | 9.75 |
ENSDART00000183987
ENSDART00000066391 |
csnk1e
|
casein kinase 1, epsilon |
| chr6_-_11792152 | 9.60 |
ENSDART00000183403
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr18_+_38749547 | 9.54 |
ENSDART00000143735
|
si:ch211-215d8.2
|
si:ch211-215d8.2 |
| chr12_-_31103187 | 9.44 |
ENSDART00000005562
ENSDART00000031408 ENSDART00000125046 ENSDART00000009237 ENSDART00000122972 ENSDART00000153068 |
tcf7l2
|
transcription factor 7 like 2 |
| chr25_+_14017609 | 9.34 |
ENSDART00000129105
ENSDART00000125733 |
chst1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr19_+_4912817 | 9.32 |
ENSDART00000101658
ENSDART00000165082 |
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr6_+_55277419 | 9.25 |
ENSDART00000083670
|
CABZ01041604.1
|
|
| chr8_+_32516160 | 9.24 |
ENSDART00000061786
|
ncs1b
|
neuronal calcium sensor 1b |
| chr8_+_54081819 | 9.21 |
ENSDART00000005857
ENSDART00000161795 |
prickle2a
|
prickle homolog 2a |
| chr2_-_32558795 | 9.20 |
ENSDART00000140026
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr13_-_18691041 | 9.14 |
ENSDART00000057867
|
sfxn3
|
sideroflexin 3 |
| chr5_+_38276582 | 9.11 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr18_-_8396192 | 9.08 |
ENSDART00000193830
|
BEND7
|
si:ch211-220f12.4 |
| chr17_+_9308425 | 8.99 |
ENSDART00000188283
ENSDART00000183311 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr2_+_49522178 | 8.98 |
ENSDART00000056254
|
stap2a
|
signal transducing adaptor family member 2a |
| chr1_-_26023678 | 8.95 |
ENSDART00000054202
|
si:ch211-145b13.5
|
si:ch211-145b13.5 |
| chr9_-_48214216 | 8.94 |
ENSDART00000012938
|
phgdh
|
phosphoglycerate dehydrogenase |
| chr5_-_66702479 | 8.88 |
ENSDART00000129197
|
mn1b
|
meningioma 1b |
| chr21_-_23475361 | 8.84 |
ENSDART00000156658
ENSDART00000157454 |
ncam1a
|
neural cell adhesion molecule 1a |
| chr10_+_26515946 | 8.81 |
ENSDART00000134276
|
synj1
|
synaptojanin 1 |
| chr8_+_47342586 | 8.71 |
ENSDART00000007624
|
plch2a
|
phospholipase C, eta 2a |
| chr2_-_34555945 | 8.70 |
ENSDART00000056671
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr24_+_4977862 | 8.68 |
ENSDART00000114537
|
zic4
|
zic family member 4 |
| chr24_-_8729716 | 8.57 |
ENSDART00000183624
|
tfap2a
|
transcription factor AP-2 alpha |
| chr11_+_25112269 | 8.56 |
ENSDART00000147546
|
ndrg3a
|
ndrg family member 3a |
| chr6_+_54888493 | 8.53 |
ENSDART00000113331
|
nav1b
|
neuron navigator 1b |
| chr16_-_34338537 | 8.50 |
ENSDART00000142223
|
hivep3b
|
human immunodeficiency virus type I enhancer binding protein 3b |
| chr1_+_15226268 | 8.38 |
ENSDART00000109911
|
hgsnat
|
heparan-alpha-glucosaminide N-acetyltransferase |
| chr23_+_24611747 | 8.36 |
ENSDART00000134978
|
nckap5l
|
NCK-associated protein 5-like |
| chr3_+_17951790 | 8.30 |
ENSDART00000164663
|
aclya
|
ATP citrate lyase a |
| chr7_-_69983948 | 8.28 |
ENSDART00000185827
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr8_-_40271264 | 8.23 |
ENSDART00000188931
|
kdm2ba
|
lysine (K)-specific demethylase 2Ba |
| chr13_-_29406534 | 8.19 |
ENSDART00000100877
|
zgc:153142
|
zgc:153142 |
| chr3_+_50312422 | 8.12 |
ENSDART00000157689
|
gas7a
|
growth arrest-specific 7a |
| chr2_-_10062575 | 8.09 |
ENSDART00000091726
|
fam78ba
|
family with sequence similarity 78, member B a |
| chr8_-_7308373 | 8.06 |
ENSDART00000132009
ENSDART00000145345 |
grip2a
|
glutamate receptor interacting protein 2a |
| chr1_-_20271138 | 8.02 |
ENSDART00000185931
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr22_-_16758438 | 8.01 |
ENSDART00000132829
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr18_+_22793743 | 7.93 |
ENSDART00000150106
|
gnao1a
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a |
| chr3_+_37197686 | 7.85 |
ENSDART00000151144
|
fmnl1a
|
formin-like 1a |
| chr25_-_518656 | 7.83 |
ENSDART00000156421
|
myo9ab
|
myosin IXAb |
| chr22_-_22416337 | 7.78 |
ENSDART00000142947
ENSDART00000089569 |
camsap2a
|
calmodulin regulated spectrin-associated protein family, member 2a |
| chr11_-_43473824 | 7.77 |
ENSDART00000179561
|
tmem63bb
|
transmembrane protein 63Bb |
| chr18_+_19419120 | 7.77 |
ENSDART00000025107
|
map2k1
|
mitogen-activated protein kinase kinase 1 |
| chr14_-_7245971 | 7.75 |
ENSDART00000108796
|
stox2b
|
storkhead box 2b |
| chr1_+_10003193 | 7.72 |
ENSDART00000162675
|
trim2b
|
tripartite motif containing 2b |
| chr10_+_44584614 | 7.71 |
ENSDART00000163523
|
sez6l
|
seizure related 6 homolog (mouse)-like |
| chr21_-_36972127 | 7.70 |
ENSDART00000100310
|
dbn1
|
drebrin 1 |
| chr13_-_10261383 | 7.69 |
ENSDART00000080808
|
six3a
|
SIX homeobox 3a |
| chr20_-_15132151 | 7.68 |
ENSDART00000063884
|
si:dkey-239i20.4
|
si:dkey-239i20.4 |
| chr14_-_2032753 | 7.68 |
ENSDART00000111335
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr4_-_69189894 | 7.62 |
ENSDART00000169596
|
si:ch211-209j12.1
|
si:ch211-209j12.1 |
| chr15_-_9272328 | 7.62 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr4_+_4803698 | 7.57 |
ENSDART00000129252
|
slc13a4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr15_-_25629046 | 7.48 |
ENSDART00000155880
|
taok1a
|
TAO kinase 1a |
| chr19_+_13099541 | 7.47 |
ENSDART00000171607
ENSDART00000165448 ENSDART00000170365 |
rims2b
|
regulating synaptic membrane exocytosis 2b |
| chr8_-_19216657 | 7.46 |
ENSDART00000135096
ENSDART00000135869 ENSDART00000145951 |
si:ch73-222f22.2
si:ch73-222f22.2
|
si:ch73-222f22.2 si:ch73-222f22.2 |
| chr5_-_38384289 | 7.46 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr2_+_45484183 | 7.43 |
ENSDART00000183490
|
si:ch211-66k16.28
|
si:ch211-66k16.28 |
| chr19_+_9455218 | 7.42 |
ENSDART00000139385
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr20_+_26939742 | 7.41 |
ENSDART00000138369
ENSDART00000062061 ENSDART00000152992 |
cdca4
|
cell division cycle associated 4 |
| chr4_+_9279784 | 7.40 |
ENSDART00000014897
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr20_-_37629084 | 7.40 |
ENSDART00000141734
|
hivep2a
|
human immunodeficiency virus type I enhancer binding protein 2a |
| chr22_-_18491813 | 7.40 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr5_+_57773222 | 7.33 |
ENSDART00000135344
|
ppp2r1ba
|
protein phosphatase 2, regulatory subunit A, beta a |
| chr13_-_23756700 | 7.28 |
ENSDART00000057612
|
rgs17
|
regulator of G protein signaling 17 |
| chr20_+_34717403 | 7.26 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr18_-_21267103 | 7.24 |
ENSDART00000136285
|
pnp6
|
purine nucleoside phosphorylase 6 |
| chr12_+_16087077 | 7.18 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr8_+_20951590 | 7.18 |
ENSDART00000138728
|
si:dkeyp-82a1.1
|
si:dkeyp-82a1.1 |
| chr19_+_27448625 | 7.17 |
ENSDART00000052352
|
ppp1r11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr10_-_25852517 | 7.17 |
ENSDART00000191551
|
trpc4a
|
transient receptor potential cation channel, subfamily C, member 4a |
| chr6_+_58698475 | 7.11 |
ENSDART00000056138
|
igsf8
|
immunoglobulin superfamily, member 8 |
| chr24_+_4978055 | 7.10 |
ENSDART00000045813
|
zic4
|
zic family member 4 |
| chr2_+_38881165 | 7.08 |
ENSDART00000141850
|
carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr20_-_48485354 | 7.04 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr9_-_10145795 | 7.04 |
ENSDART00000004745
ENSDART00000143295 |
hnmt
|
histamine N-methyltransferase |
| chr19_-_28130658 | 7.02 |
ENSDART00000079114
|
irx1b
|
iroquois homeobox 1b |
| chr3_-_42086577 | 6.95 |
ENSDART00000083111
ENSDART00000187312 |
ttyh3a
|
tweety family member 3a |
| chr11_-_39044595 | 6.93 |
ENSDART00000065461
|
cldn19
|
claudin 19 |
| chr2_-_36819624 | 6.92 |
ENSDART00000140844
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr15_-_26636826 | 6.89 |
ENSDART00000087632
|
slc47a4
|
solute carrier family 47 (multidrug and toxin extrusion), member 4 |
| chr10_+_13279079 | 6.86 |
ENSDART00000135082
|
tmem267
|
transmembrane protein 267 |
| chr13_+_3667230 | 6.85 |
ENSDART00000131553
ENSDART00000189841 ENSDART00000183554 ENSDART00000018737 |
qkib
|
QKI, KH domain containing, RNA binding b |
| chr7_+_1521834 | 6.81 |
ENSDART00000174007
|
si:cabz01102082.1
|
si:cabz01102082.1 |
| chr24_+_5840258 | 6.80 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr2_-_37043905 | 6.79 |
ENSDART00000056514
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr13_-_24218795 | 6.76 |
ENSDART00000136217
|
galnt2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 |
| chr9_+_17787864 | 6.73 |
ENSDART00000013111
|
dgkh
|
diacylglycerol kinase, eta |
| chr23_+_21455152 | 6.66 |
ENSDART00000158511
ENSDART00000161321 ENSDART00000160731 ENSDART00000137573 |
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr14_+_33722950 | 6.64 |
ENSDART00000075312
|
apln
|
apelin |
| chr14_-_33265068 | 6.63 |
ENSDART00000147038
|
gdpd2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr8_+_16758304 | 6.62 |
ENSDART00000133514
|
elovl7a
|
ELOVL fatty acid elongase 7a |
| chr14_+_33525196 | 6.57 |
ENSDART00000085335
|
zdhhc9
|
zinc finger, DHHC-type containing 9 |
| chr9_-_16877456 | 6.55 |
ENSDART00000161105
ENSDART00000160869 |
fbxl3a
|
F-box and leucine-rich repeat protein 3a |
Network of associatons between targets according to the STRING database.
First level regulatory network of gfi1aa+gfi1ab
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.8 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 3.8 | 15.0 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 3.6 | 32.3 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 3.4 | 13.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 3.0 | 8.9 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 2.9 | 8.8 | GO:0021961 | posterior commissure morphogenesis(GO:0021961) |
| 2.6 | 18.5 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 2.6 | 56.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 2.4 | 9.4 | GO:0010226 | response to lithium ion(GO:0010226) |
| 2.3 | 7.0 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 2.3 | 6.8 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 2.2 | 11.0 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 2.2 | 15.3 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 2.1 | 14.7 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 2.0 | 22.4 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 2.0 | 20.2 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 2.0 | 35.2 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 1.9 | 24.4 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 1.9 | 37.4 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 1.8 | 5.5 | GO:0015808 | L-alanine transport(GO:0015808) |
| 1.8 | 25.6 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 1.8 | 5.3 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 1.8 | 5.3 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 1.8 | 5.3 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 1.8 | 5.3 | GO:0036315 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 1.7 | 16.8 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 1.6 | 4.8 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 1.6 | 7.8 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 1.4 | 11.6 | GO:0046323 | glucose import(GO:0046323) |
| 1.4 | 18.4 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 1.4 | 9.9 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 1.3 | 4.0 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 1.3 | 20.9 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 1.3 | 5.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 1.3 | 6.4 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 1.3 | 6.4 | GO:0007412 | axon target recognition(GO:0007412) |
| 1.3 | 5.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 1.3 | 12.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 1.2 | 12.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 1.2 | 4.7 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 1.2 | 12.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 1.2 | 4.6 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 1.0 | 45.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 1.0 | 9.2 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 1.0 | 5.0 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 1.0 | 14.8 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 1.0 | 28.5 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.9 | 13.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.9 | 7.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.9 | 2.6 | GO:2000252 | negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.9 | 5.2 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.8 | 11.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.8 | 5.8 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.8 | 5.8 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.8 | 5.8 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.8 | 8.9 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.8 | 11.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.8 | 5.6 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.8 | 4.7 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.8 | 2.4 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.8 | 12.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.8 | 11.6 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.8 | 2.3 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.8 | 2.3 | GO:1904869 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.8 | 6.1 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.7 | 7.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.7 | 7.4 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) reactive nitrogen species metabolic process(GO:2001057) |
| 0.7 | 2.2 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.7 | 10.9 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.7 | 2.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.7 | 11.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.7 | 2.1 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.7 | 15.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.7 | 31.9 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.7 | 2.0 | GO:0039529 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.7 | 2.0 | GO:0051039 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.6 | 9.4 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.6 | 6.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.6 | 7.4 | GO:0001964 | startle response(GO:0001964) |
| 0.6 | 3.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.6 | 19.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.6 | 4.3 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.6 | 37.0 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.6 | 4.2 | GO:0003321 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.6 | 13.0 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.6 | 3.5 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.6 | 9.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.6 | 12.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.6 | 2.8 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.6 | 3.3 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.5 | 2.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.5 | 14.0 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.5 | 12.6 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.5 | 9.4 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.5 | 5.7 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.5 | 19.9 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.5 | 1.5 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.5 | 2.0 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.5 | 4.9 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.5 | 3.0 | GO:0050795 | regulation of behavior(GO:0050795) |
| 0.5 | 3.4 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.5 | 3.9 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.5 | 3.8 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.5 | 19.5 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.5 | 3.3 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.5 | 6.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.5 | 1.4 | GO:1903673 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.5 | 13.1 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.4 | 2.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.4 | 1.3 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.4 | 3.7 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.4 | 2.9 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.4 | 4.4 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.4 | 8.4 | GO:0043153 | entrainment of circadian clock(GO:0009649) entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.4 | 2.8 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.4 | 6.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.4 | 21.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.4 | 2.7 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.4 | 5.0 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.4 | 2.3 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.4 | 1.5 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.4 | 2.6 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.4 | 14.2 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.4 | 6.4 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.3 | 1.4 | GO:0072314 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.3 | 3.8 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.3 | 8.2 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.3 | 6.1 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.3 | 0.6 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.3 | 39.6 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.3 | 5.8 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.3 | 2.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.3 | 6.6 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.3 | 3.5 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 12.3 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.3 | 4.1 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.3 | 9.9 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.3 | 0.9 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.3 | 0.6 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.3 | 2.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.3 | 11.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.3 | 10.6 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.3 | 2.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.3 | 1.9 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.3 | 1.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 | 0.8 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.3 | 1.9 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.3 | 0.8 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.3 | 3.4 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.2 | 4.1 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.2 | 6.8 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.2 | 22.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.2 | 8.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 2.3 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.2 | 3.4 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.2 | 24.7 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.2 | 10.4 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.2 | 3.6 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.2 | 6.8 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.2 | 3.8 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.2 | 2.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.2 | 14.9 | GO:0051170 | nuclear import(GO:0051170) |
| 0.2 | 3.7 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.2 | 4.5 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.2 | 7.7 | GO:0001764 | neuron migration(GO:0001764) |
| 0.2 | 3.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.2 | 19.0 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.2 | 3.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.2 | 6.2 | GO:0001878 | response to yeast(GO:0001878) |
| 0.2 | 26.1 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.2 | 2.9 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.2 | 12.6 | GO:1990542 | mitochondrial transmembrane transport(GO:1990542) |
| 0.2 | 1.9 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.2 | 1.6 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.2 | 4.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 1.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 3.9 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.1 | 0.6 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 1.2 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 4.0 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 3.2 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.1 | 16.0 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.1 | 2.5 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.1 | 3.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 24.5 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 3.6 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.1 | 1.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 3.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 5.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 5.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 5.4 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.1 | 0.6 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 2.9 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.1 | 3.0 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 2.8 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.4 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 1.1 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 8.4 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 4.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 2.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 0.3 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA transport(GO:0051031) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.7 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 4.9 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 1.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 12.6 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.1 | 5.7 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 0.8 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 3.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 3.0 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.1 | 0.5 | GO:0035475 | angioblast cell migration involved in selective angioblast sprouting(GO:0035475) |
| 0.1 | 9.2 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.1 | 3.1 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 0.6 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.1 | 0.3 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.1 | 3.5 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 1.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 2.0 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 1.0 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 0.5 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 2.3 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 1.2 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.1 | 4.4 | GO:0006821 | chloride transport(GO:0006821) |
| 0.1 | 1.8 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.1 | 4.4 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 2.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 1.3 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.1 | 0.4 | GO:1905066 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 2.4 | GO:0061572 | actin filament bundle organization(GO:0061572) |
| 0.0 | 0.7 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.8 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 2.5 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.0 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 1.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 1.1 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 3.2 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 1.1 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 0.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.8 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 21.3 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 3.2 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.2 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 1.6 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.3 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 2.5 | GO:0055080 | cation homeostasis(GO:0055080) |
| 0.0 | 0.4 | GO:0072666 | protein targeting to vacuole(GO:0006623) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 | 1.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.5 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.4 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 22.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 4.7 | 18.6 | GO:0043291 | RAVE complex(GO:0043291) |
| 2.6 | 15.8 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 2.4 | 9.4 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 2.3 | 67.7 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 2.2 | 37.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 2.2 | 6.5 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 1.8 | 5.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 1.5 | 12.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 1.4 | 4.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 1.4 | 5.6 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 1.4 | 4.1 | GO:1990745 | EARP complex(GO:1990745) |
| 1.3 | 5.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 1.2 | 8.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 1.2 | 5.9 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 1.0 | 49.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 1.0 | 43.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 1.0 | 22.6 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 1.0 | 30.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.9 | 2.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.9 | 9.4 | GO:1904949 | ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.9 | 12.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.8 | 12.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.8 | 7.8 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.8 | 3.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.7 | 4.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.7 | 2.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.7 | 4.7 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.7 | 21.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.6 | 1.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.6 | 11.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.6 | 19.7 | GO:0043679 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.6 | 9.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.6 | 2.8 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.5 | 3.5 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.5 | 10.9 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.5 | 8.3 | GO:0035371 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.5 | 3.6 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.4 | 4.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.4 | 8.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.4 | 6.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.4 | 3.7 | GO:0089701 | U2AF(GO:0089701) |
| 0.4 | 34.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.4 | 11.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.4 | 13.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.4 | 19.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.4 | 3.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.4 | 13.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.3 | 2.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 4.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.3 | 2.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.3 | 3.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.3 | 4.0 | GO:0070187 | telosome(GO:0070187) |
| 0.3 | 14.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.3 | 2.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 3.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 8.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 4.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.2 | 51.3 | GO:0030424 | axon(GO:0030424) |
| 0.2 | 5.0 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.2 | 4.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 5.5 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.2 | 3.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 14.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 6.8 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.2 | 0.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 7.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 1.4 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 12.6 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.2 | 2.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 7.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 14.0 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.2 | 2.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 3.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 2.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 25.8 | GO:0036477 | dendrite(GO:0030425) somatodendritic compartment(GO:0036477) |
| 0.1 | 0.4 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.1 | 0.7 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 10.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 2.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 37.3 | GO:0043005 | neuron projection(GO:0043005) |
| 0.1 | 11.9 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 0.6 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 8.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 5.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 4.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 2.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 23.4 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.1 | 1.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.6 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 39.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 0.4 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 0.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 5.9 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.1 | 22.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 26.7 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.1 | 2.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.2 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 5.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 1.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 1.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 13.8 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 3.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.1 | GO:0060091 | radial spoke(GO:0001534) kinocilium(GO:0060091) |
| 0.0 | 0.7 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 38.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.5 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 13.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 2.2 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.0 | 0.8 | GO:0045495 | pole plasm(GO:0045495) |
| 0.0 | 0.5 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 38.7 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 6.1 | 18.4 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 5.7 | 85.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 3.8 | 15.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 2.9 | 35.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 2.8 | 41.6 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 2.5 | 19.7 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 2.4 | 14.6 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 2.3 | 16.3 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 2.2 | 6.5 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 2.1 | 6.4 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 2.0 | 5.9 | GO:0046978 | TAP1 binding(GO:0046978) |
| 1.8 | 5.5 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 1.8 | 37.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 1.7 | 5.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 1.7 | 10.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.6 | 11.3 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 1.6 | 14.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 1.6 | 4.7 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 1.5 | 10.8 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 1.4 | 18.4 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 1.4 | 5.6 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 1.3 | 18.0 | GO:0034596 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) |
| 1.3 | 17.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 1.2 | 9.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 1.2 | 10.8 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 1.1 | 8.9 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 1.1 | 9.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.1 | 3.2 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 1.1 | 5.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 1.1 | 7.4 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 1.0 | 6.3 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 1.0 | 8.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 1.0 | 9.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 1.0 | 12.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.9 | 27.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.9 | 16.8 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.9 | 5.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.9 | 22.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.9 | 2.6 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.9 | 7.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.9 | 28.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.9 | 13.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.9 | 7.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.8 | 48.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.8 | 7.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.8 | 21.0 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.8 | 7.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.8 | 25.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.8 | 5.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.7 | 6.0 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.7 | 7.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.7 | 12.6 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.7 | 29.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.7 | 6.5 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.7 | 6.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.7 | 8.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.7 | 4.6 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.7 | 3.9 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.7 | 15.0 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.6 | 15.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.6 | 8.6 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.6 | 10.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.6 | 15.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.6 | 34.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.6 | 2.8 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.5 | 2.2 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.5 | 11.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.5 | 5.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.5 | 12.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.5 | 4.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.5 | 3.6 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.5 | 4.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.5 | 3.1 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.5 | 7.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.5 | 3.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.5 | 9.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.5 | 5.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.5 | 10.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.5 | 2.3 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.4 | 6.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.4 | 5.2 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.4 | 6.7 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.4 | 12.8 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.4 | 36.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.4 | 10.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.4 | 1.5 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.4 | 7.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.4 | 3.7 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.3 | 11.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.3 | 2.7 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.3 | 7.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.3 | 0.9 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.3 | 2.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.3 | 6.6 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 12.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.3 | 6.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.3 | 11.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.3 | 4.6 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.3 | 1.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.3 | 2.5 | GO:0016918 | retinal binding(GO:0016918) |
| 0.2 | 18.3 | GO:0051287 | NAD binding(GO:0051287) |
| 0.2 | 7.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 1.0 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.2 | 5.7 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.2 | 4.8 | GO:0048038 | quinone binding(GO:0048038) |
| 0.2 | 1.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 0.7 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.2 | 10.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 1.1 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.2 | 1.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 7.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 2.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 3.9 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.2 | 1.9 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.2 | 1.5 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.2 | 11.0 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.2 | 8.2 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.2 | 18.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.2 | 9.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 1.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 1.0 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.2 | 1.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 14.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 16.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.2 | 1.7 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.2 | 3.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.2 | 2.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.2 | 10.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 0.8 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 3.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 4.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 9.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.2 | 0.6 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 2.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 3.6 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 0.4 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.1 | 3.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 6.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 3.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 4.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 3.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 18.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 2.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 2.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 3.1 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 0.6 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 9.0 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 1.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.1 | GO:0005487 | nuclear export signal receptor activity(GO:0005049) nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 14.4 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.1 | 2.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 5.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 14.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 4.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 18.8 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 15.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 5.3 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 2.3 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 3.4 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.1 | 2.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 4.4 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 4.4 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 2.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 1.5 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 1.6 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 5.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 5.6 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 1.2 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.1 | 1.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.6 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.4 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 6.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.8 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 1.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 14.4 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.6 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 14.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.1 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 2.5 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 16.6 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 2.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 5.2 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 2.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.8 | 5.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.7 | 11.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.5 | 22.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.4 | 2.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.4 | 12.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.4 | 7.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.4 | 9.5 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.3 | 2.4 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.2 | 11.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 12.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 4.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 8.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.2 | 4.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 3.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 8.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 4.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 5.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.2 | 5.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 3.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 4.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 2.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 46.6 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 1.4 | 12.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 1.4 | 12.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 1.3 | 7.8 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.9 | 8.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.8 | 10.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.8 | 21.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.7 | 4.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.7 | 5.8 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.7 | 7.8 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.7 | 5.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.7 | 9.2 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.6 | 8.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.6 | 4.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.6 | 4.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.6 | 13.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.5 | 10.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.5 | 4.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.4 | 2.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.4 | 4.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.4 | 4.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.4 | 5.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.4 | 6.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.3 | 3.8 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.3 | 16.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.3 | 1.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 5.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.3 | 6.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.3 | 2.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.3 | 15.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.3 | 5.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 17.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 7.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 6.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 3.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 1.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 9.8 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.2 | 1.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 2.6 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.2 | 2.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 1.3 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 4.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 3.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 5.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 3.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 4.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 0.7 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 2.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |