Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
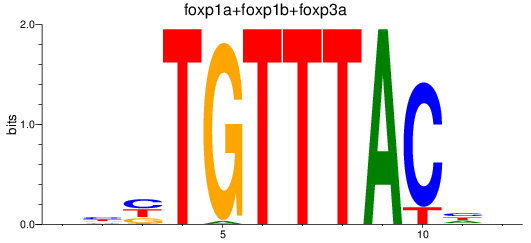
Results for foxp1a+foxp1b+foxp3a_foxg1b_foxp2_foxd1+foxd7_foxo1a+foxo1b
Z-value: 2.80
Transcription factors associated with foxp1a+foxp1b+foxp3a_foxg1b_foxp2_foxd1+foxd7_foxo1a+foxo1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxp1a
|
ENSDARG00000004843 | forkhead box P1a |
|
foxp1b
|
ENSDARG00000014181 | forkhead box P1b |
|
foxp3a
|
ENSDARG00000055750 | forkhead box P3a |
|
foxg1b
|
ENSDARG00000032705 | forkhead box G1b |
|
foxp2
|
ENSDARG00000005453 | forkhead box P2 |
|
foxd1
|
ENSDARG00000029179 | forkhead box D1 |
|
foxd7
|
ENSDARG00000079699 | zgc |
|
foxo1b
|
ENSDARG00000061549 | forkhead box O1 b |
|
foxo1a
|
ENSDARG00000099555 | forkhead box O1 a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxp2 | dr11_v1_chr4_-_6567355_6567503 | 0.81 | 4.7e-23 | Click! |
| foxg1b | dr11_v1_chr11_+_43114108_43114108 | 0.62 | 2.0e-11 | Click! |
| foxd1 | dr11_v1_chr5_+_34997763_34997763 | 0.28 | 6.6e-03 | Click! |
| foxp3a | dr11_v1_chr8_-_25728628_25728628 | 0.27 | 9.1e-03 | Click! |
| foxp1a | dr11_v1_chr23_-_10723009_10723009 | 0.26 | 9.9e-03 | Click! |
| foxp1b | dr11_v1_chr6_-_43677125_43677125 | 0.25 | 1.6e-02 | Click! |
| foxo1a | dr11_v1_chr15_+_3284684_3284684 | -0.20 | 5.9e-02 | Click! |
| foxo1b | dr11_v1_chr10_+_31809226_31809226 | -0.15 | 1.5e-01 | Click! |
| zgc:162612 | dr11_v1_chr3_-_26109322_26109322 | -0.04 | 7.2e-01 | Click! |
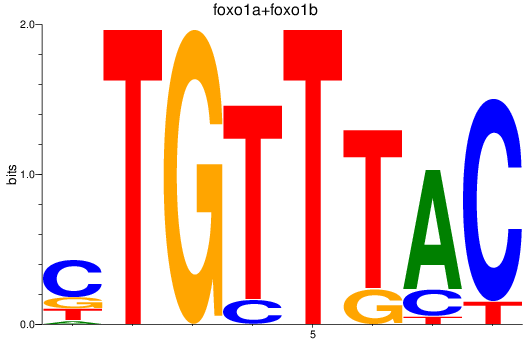
Activity profile of foxp1a+foxp1b+foxp3a_foxg1b_foxp2_foxd1+foxd7_foxo1a+foxo1b motif
Sorted Z-values of foxp1a+foxp1b+foxp3a_foxg1b_foxp2_foxd1+foxd7_foxo1a+foxo1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_48460239 | 56.13 |
ENSDART00000052113
|
lingo1b
|
leucine rich repeat and Ig domain containing 1b |
| chr19_+_10396042 | 46.95 |
ENSDART00000028048
ENSDART00000151735 |
necap1
|
NECAP endocytosis associated 1 |
| chr14_-_26177156 | 45.57 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr3_-_21280373 | 38.74 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr14_-_36378494 | 38.59 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr14_-_7885707 | 36.02 |
ENSDART00000029981
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr20_+_27393668 | 35.73 |
ENSDART00000005473
|
tmem179
|
transmembrane protein 179 |
| chr23_-_19953089 | 35.43 |
ENSDART00000153828
|
atp2b3b
|
ATPase plasma membrane Ca2+ transporting 3b |
| chr12_+_25640480 | 33.85 |
ENSDART00000105608
|
prkcea
|
protein kinase C, epsilon a |
| chr16_-_43025885 | 33.52 |
ENSDART00000193146
ENSDART00000157302 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr2_-_38125657 | 32.73 |
ENSDART00000143433
|
cbln12
|
cerebellin 12 |
| chr21_-_29100110 | 31.11 |
ENSDART00000142598
|
timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr6_+_36942966 | 31.08 |
ENSDART00000028895
|
negr1
|
neuronal growth regulator 1 |
| chr17_-_6738538 | 30.86 |
ENSDART00000157125
|
vsnl1b
|
visinin-like 1b |
| chr2_-_8017579 | 29.54 |
ENSDART00000040209
|
ephb3a
|
eph receptor B3a |
| chr14_-_34044369 | 28.72 |
ENSDART00000149396
ENSDART00000123607 ENSDART00000190746 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr25_-_19433244 | 28.18 |
ENSDART00000154778
|
map1ab
|
microtubule-associated protein 1Ab |
| chr19_-_5103313 | 27.73 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr6_-_32999646 | 26.89 |
ENSDART00000159510
|
adcyap1r1b
|
adenylate cyclase activating polypeptide 1b (pituitary) receptor type I |
| chr15_-_33933790 | 25.81 |
ENSDART00000165162
ENSDART00000182258 ENSDART00000183240 |
mag
|
myelin associated glycoprotein |
| chr25_-_15049694 | 25.78 |
ENSDART00000162485
ENSDART00000164384 ENSDART00000165632 ENSDART00000159490 |
pax6a
|
paired box 6a |
| chr23_-_29502287 | 25.63 |
ENSDART00000141075
ENSDART00000053807 |
kif1b
|
kinesin family member 1B |
| chr1_+_53954230 | 24.72 |
ENSDART00000037729
ENSDART00000159900 |
ccsapa
|
centriole, cilia and spindle-associated protein a |
| chr21_-_25522906 | 24.70 |
ENSDART00000110923
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr6_-_40744720 | 24.70 |
ENSDART00000154916
ENSDART00000186922 |
p4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr17_-_40397752 | 24.62 |
ENSDART00000178483
|
BX548062.1
|
|
| chr20_-_19590378 | 24.50 |
ENSDART00000152588
|
baalcb
|
brain and acute leukemia, cytoplasmic b |
| chr21_+_11684830 | 24.42 |
ENSDART00000147473
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr1_-_44901163 | 23.96 |
ENSDART00000145354
|
tcirg1a
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3a |
| chr12_+_31729075 | 23.78 |
ENSDART00000152973
|
RNF157
|
si:dkey-49c17.3 |
| chr13_+_1100197 | 23.64 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr21_+_5209716 | 23.48 |
ENSDART00000102539
ENSDART00000053148 ENSDART00000102536 |
st8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr23_+_16620801 | 23.37 |
ENSDART00000189859
ENSDART00000184578 |
snphb
|
syntaphilin b |
| chr4_+_8168514 | 23.28 |
ENSDART00000150830
|
ninj2
|
ninjurin 2 |
| chr11_+_25112269 | 23.17 |
ENSDART00000147546
|
ndrg3a
|
ndrg family member 3a |
| chr22_+_27090136 | 23.02 |
ENSDART00000136770
|
si:dkey-246e1.3
|
si:dkey-246e1.3 |
| chr10_+_33895315 | 23.01 |
ENSDART00000142881
|
fryb
|
furry homolog b (Drosophila) |
| chr7_+_30787903 | 23.00 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr1_+_16127825 | 22.83 |
ENSDART00000122503
|
tusc3
|
tumor suppressor candidate 3 |
| chr6_+_21095918 | 22.56 |
ENSDART00000167225
|
spega
|
SPEG complex locus a |
| chr6_+_38427570 | 22.56 |
ENSDART00000170612
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr21_+_5209476 | 22.28 |
ENSDART00000146400
|
st8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr24_-_4973765 | 22.22 |
ENSDART00000127597
|
zic1
|
zic family member 1 (odd-paired homolog, Drosophila) |
| chr9_+_52492639 | 21.65 |
ENSDART00000078939
|
march4
|
membrane-associated ring finger (C3HC4) 4 |
| chr15_+_18130109 | 21.49 |
ENSDART00000078723
|
dixdc1a
|
DIX domain containing 1a |
| chr15_+_32249062 | 21.24 |
ENSDART00000133867
ENSDART00000152545 ENSDART00000082315 ENSDART00000152513 ENSDART00000152139 |
arfip2a
|
ADP-ribosylation factor interacting protein 2a |
| chr3_-_51912019 | 20.66 |
ENSDART00000149914
|
aatka
|
apoptosis-associated tyrosine kinase a |
| chr21_-_42097736 | 20.63 |
ENSDART00000100000
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr18_-_22094102 | 20.58 |
ENSDART00000100904
|
pard6a
|
par-6 family cell polarity regulator alpha |
| chr17_-_26911852 | 20.19 |
ENSDART00000045842
|
rcan3
|
regulator of calcineurin 3 |
| chr21_+_11685009 | 20.14 |
ENSDART00000014668
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr19_-_5103141 | 20.06 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr2_+_26240631 | 20.03 |
ENSDART00000129895
|
palm1b
|
paralemmin 1b |
| chr23_-_29505463 | 19.96 |
ENSDART00000050915
|
kif1b
|
kinesin family member 1B |
| chr18_+_17428506 | 19.66 |
ENSDART00000100223
|
zgc:91860
|
zgc:91860 |
| chr11_-_3552067 | 19.65 |
ENSDART00000163656
|
CAMK2N1
|
si:dkey-33m11.6 |
| chr7_+_36898850 | 19.59 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr5_-_23317477 | 19.50 |
ENSDART00000090171
|
nlgn3b
|
neuroligin 3b |
| chr1_+_25801648 | 19.29 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr20_+_30445971 | 19.18 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr3_+_32403758 | 19.12 |
ENSDART00000156982
|
si:ch211-195b15.8
|
si:ch211-195b15.8 |
| chr12_-_4781801 | 19.08 |
ENSDART00000167490
ENSDART00000121718 |
mapta
|
microtubule-associated protein tau a |
| chr11_-_8126223 | 18.47 |
ENSDART00000091617
ENSDART00000192391 ENSDART00000101561 |
ttll7
|
tubulin tyrosine ligase-like family, member 7 |
| chr11_+_6819050 | 17.99 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr5_-_40190949 | 17.75 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr15_-_18574716 | 17.73 |
ENSDART00000142010
ENSDART00000019006 |
ncam1b
|
neural cell adhesion molecule 1b |
| chr7_-_38612230 | 17.35 |
ENSDART00000173678
|
c1qtnf4
|
C1q and TNF related 4 |
| chr24_-_6678640 | 17.11 |
ENSDART00000042478
|
enkur
|
enkurin, TRPC channel interacting protein |
| chr13_-_11378355 | 17.09 |
ENSDART00000164566
|
akt3a
|
v-akt murine thymoma viral oncogene homolog 3a |
| chr3_-_28258462 | 17.09 |
ENSDART00000191573
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr15_+_8043751 | 16.91 |
ENSDART00000193701
|
cadm2b
|
cell adhesion molecule 2b |
| chr7_-_35710263 | 16.83 |
ENSDART00000043857
|
irx5a
|
iroquois homeobox 5a |
| chr5_+_52625975 | 16.77 |
ENSDART00000170341
ENSDART00000168317 |
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr5_-_20879527 | 16.65 |
ENSDART00000134697
|
pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr7_-_58098814 | 16.56 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr23_-_36449111 | 16.09 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr12_-_7607114 | 16.08 |
ENSDART00000158095
|
slc16a9b
|
solute carrier family 16, member 9b |
| chr11_+_11120532 | 15.92 |
ENSDART00000026135
ENSDART00000189872 |
ly75
|
lymphocyte antigen 75 |
| chr4_-_14207471 | 15.91 |
ENSDART00000015134
|
twf1b
|
twinfilin actin-binding protein 1b |
| chr19_-_7358184 | 15.90 |
ENSDART00000092379
|
oxr1b
|
oxidation resistance 1b |
| chr3_-_60142530 | 15.79 |
ENSDART00000153247
|
si:ch211-120g10.1
|
si:ch211-120g10.1 |
| chr13_+_22280983 | 15.64 |
ENSDART00000173258
ENSDART00000173379 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr21_-_41305748 | 15.54 |
ENSDART00000170457
|
nsg2
|
neuronal vesicle trafficking associated 2 |
| chr14_-_48939560 | 15.53 |
ENSDART00000021736
|
scocb
|
short coiled-coil protein b |
| chr7_-_33023404 | 15.29 |
ENSDART00000052383
|
cd81a
|
CD81 molecule a |
| chr20_-_16849306 | 15.28 |
ENSDART00000131395
ENSDART00000027582 |
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr12_+_28367557 | 15.26 |
ENSDART00000066294
|
cdk5r1b
|
cyclin-dependent kinase 5, regulatory subunit 1b (p35) |
| chr11_-_24191928 | 15.25 |
ENSDART00000136827
|
sox12
|
SRY (sex determining region Y)-box 12 |
| chr14_-_4273396 | 15.00 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr9_-_3149896 | 14.93 |
ENSDART00000020861
|
pdk1
|
pyruvate dehydrogenase kinase, isozyme 1 |
| chr18_-_1228688 | 14.66 |
ENSDART00000064403
|
nptnb
|
neuroplastin b |
| chr14_+_9421510 | 14.58 |
ENSDART00000123652
|
hmgn6
|
high mobility group nucleosome binding domain 6 |
| chr21_+_17301790 | 14.58 |
ENSDART00000145057
|
tsc1b
|
TSC complex subunit 1b |
| chr12_+_22404108 | 14.43 |
ENSDART00000153055
|
hdlbpb
|
high density lipoprotein binding protein b |
| chr2_+_22795494 | 14.40 |
ENSDART00000042255
|
rab6bb
|
RAB6B, member RAS oncogene family b |
| chr13_-_25842074 | 14.36 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr8_-_18262149 | 13.95 |
ENSDART00000143000
|
rnf220b
|
ring finger protein 220b |
| chr7_+_28724919 | 13.90 |
ENSDART00000011324
|
ccdc102a
|
coiled-coil domain containing 102A |
| chr3_-_26191960 | 13.71 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr2_-_12243213 | 13.61 |
ENSDART00000113081
|
gpr158b
|
G protein-coupled receptor 158b |
| chr6_-_41138854 | 13.58 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr7_-_37555208 | 13.52 |
ENSDART00000148905
ENSDART00000150229 |
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr24_-_6647275 | 13.33 |
ENSDART00000161494
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr24_-_21498802 | 13.20 |
ENSDART00000181235
ENSDART00000153695 |
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr9_+_17348745 | 13.19 |
ENSDART00000147488
|
slain1a
|
SLAIN motif family, member 1a |
| chr13_-_12006007 | 13.09 |
ENSDART00000111438
|
mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr7_-_31941670 | 13.03 |
ENSDART00000180929
ENSDART00000075389 |
bdnf
|
brain-derived neurotrophic factor |
| chr1_+_51312752 | 13.02 |
ENSDART00000063938
|
mast1a
|
microtubule associated serine/threonine kinase 1a |
| chr20_-_34801181 | 12.95 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr6_-_30658755 | 12.83 |
ENSDART00000065215
ENSDART00000181302 |
lurap1
|
leucine rich adaptor protein 1 |
| chr13_-_12005429 | 12.64 |
ENSDART00000180302
|
mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr2_-_24462277 | 12.55 |
ENSDART00000033922
|
kcnn1a
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1a |
| chr14_-_21219659 | 12.53 |
ENSDART00000089867
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr7_-_30082931 | 12.53 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr9_+_41459759 | 12.45 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr7_-_31618166 | 12.44 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr24_+_19210001 | 12.40 |
ENSDART00000179373
ENSDART00000139299 |
zgc:162928
|
zgc:162928 |
| chr9_+_33154841 | 12.21 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr11_-_29563437 | 12.18 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr21_+_25071805 | 12.16 |
ENSDART00000078651
|
dixdc1b
|
DIX domain containing 1b |
| chr7_+_69470142 | 12.16 |
ENSDART00000073861
|
gabarapb
|
GABA(A) receptor-associated protein b |
| chr13_+_28612313 | 12.07 |
ENSDART00000077383
|
borcs7
|
BLOC-1 related complex subunit 7 |
| chr11_+_7580079 | 12.03 |
ENSDART00000091550
ENSDART00000193223 ENSDART00000193386 |
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr12_+_31729498 | 12.02 |
ENSDART00000188546
ENSDART00000182562 ENSDART00000186147 |
RNF157
|
si:dkey-49c17.3 |
| chr5_+_61799629 | 11.99 |
ENSDART00000113508
|
hnrnpul1l
|
heterogeneous nuclear ribonucleoprotein U-like 1 like |
| chr20_+_22666548 | 11.87 |
ENSDART00000147520
|
lnx1
|
ligand of numb-protein X 1 |
| chr3_-_13147310 | 11.86 |
ENSDART00000160840
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr2_+_26240339 | 11.84 |
ENSDART00000191006
|
palm1b
|
paralemmin 1b |
| chr10_+_22890791 | 11.76 |
ENSDART00000176011
|
arrb2a
|
arrestin, beta 2a |
| chr7_+_69470442 | 11.73 |
ENSDART00000189593
|
gabarapb
|
GABA(A) receptor-associated protein b |
| chr3_+_32933663 | 11.70 |
ENSDART00000112742
|
nbr1b
|
neighbor of brca1 gene 1b |
| chr5_-_66749535 | 11.68 |
ENSDART00000132183
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr2_-_5942115 | 11.58 |
ENSDART00000154489
|
tmem125b
|
transmembrane protein 125b |
| chr17_-_14966384 | 11.48 |
ENSDART00000105064
|
txndc16
|
thioredoxin domain containing 16 |
| chr18_-_5781922 | 11.45 |
ENSDART00000128722
|
RGS9BP
|
si:ch73-167i17.6 |
| chr10_+_22891126 | 11.45 |
ENSDART00000057291
|
arrb2a
|
arrestin, beta 2a |
| chr9_+_38163876 | 11.42 |
ENSDART00000137955
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr14_-_14746051 | 11.36 |
ENSDART00000163199
ENSDART00000170623 ENSDART00000158629 ENSDART00000171581 |
ogt.1
|
O-linked N-acetylglucosamine (GlcNAc) transferase, tandem duplicate 1 |
| chr18_-_13121983 | 11.32 |
ENSDART00000092648
|
rxylt1
|
ribitol xylosyltransferase 1 |
| chr25_-_25434479 | 11.32 |
ENSDART00000171589
|
hrasa
|
v-Ha-ras Harvey rat sarcoma viral oncogene homolog a |
| chr14_+_17197132 | 11.26 |
ENSDART00000054598
|
rtn4rl2b
|
reticulon 4 receptor-like 2b |
| chr4_-_5764255 | 11.23 |
ENSDART00000113864
|
faxca
|
failed axon connections homolog a |
| chr6_+_38427357 | 11.17 |
ENSDART00000148678
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr23_-_27608257 | 11.10 |
ENSDART00000026314
|
phf8
|
PHD finger protein 8 |
| chr22_-_21046654 | 10.99 |
ENSDART00000064902
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr8_+_23639124 | 10.99 |
ENSDART00000083108
|
nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr17_+_18117029 | 10.98 |
ENSDART00000154646
ENSDART00000179739 |
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr3_-_13146631 | 10.98 |
ENSDART00000172460
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr7_-_24828296 | 10.96 |
ENSDART00000138193
|
otub1b
|
OTU deubiquitinase, ubiquitin aldehyde binding 1b |
| chr25_+_3677650 | 10.79 |
ENSDART00000154348
|
prnprs3
|
prion protein, related sequence 3 |
| chr20_+_10166297 | 10.77 |
ENSDART00000141877
|
kcnk10a
|
potassium channel, subfamily K, member 10a |
| chr1_+_34763539 | 10.76 |
ENSDART00000077725
ENSDART00000113808 |
zgc:172122
|
zgc:172122 |
| chr10_-_32890617 | 10.74 |
ENSDART00000134922
|
kctd7
|
potassium channel tetramerization domain containing 7 |
| chr20_+_40457599 | 10.70 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr6_-_21616659 | 10.67 |
ENSDART00000074256
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr25_+_388258 | 10.66 |
ENSDART00000166834
|
rfx7b
|
regulatory factor X7b |
| chr18_-_14941840 | 10.60 |
ENSDART00000091729
|
mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr7_+_38278860 | 10.56 |
ENSDART00000016265
|
lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr22_-_25033105 | 10.52 |
ENSDART00000124220
|
nptxrb
|
neuronal pentraxin receptor b |
| chr16_-_21047872 | 10.47 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr3_+_33340939 | 10.39 |
ENSDART00000128786
|
pyya
|
peptide YYa |
| chr11_+_18873619 | 10.35 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr3_+_26044969 | 10.29 |
ENSDART00000133523
|
hmgxb4a
|
HMG box domain containing 4a |
| chr1_-_8917902 | 10.28 |
ENSDART00000137900
|
grin2ab
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A, b |
| chr14_+_49382180 | 10.25 |
ENSDART00000158329
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr14_-_17599452 | 10.23 |
ENSDART00000080042
|
rab33a
|
RAB33A, member RAS oncogene family |
| chr16_-_28856112 | 10.21 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr5_-_25236340 | 10.19 |
ENSDART00000162774
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr1_-_40994259 | 10.17 |
ENSDART00000101562
|
adra2c
|
adrenoceptor alpha 2C |
| chr11_+_16153207 | 10.16 |
ENSDART00000192356
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr12_+_32729470 | 10.16 |
ENSDART00000175712
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr25_+_13731542 | 10.09 |
ENSDART00000161012
|
ccdc135
|
coiled-coil domain containing 135 |
| chr15_-_33896159 | 10.08 |
ENSDART00000159791
|
mag
|
myelin associated glycoprotein |
| chr15_+_26933196 | 10.02 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr23_-_30787932 | 9.99 |
ENSDART00000135771
|
myt1a
|
myelin transcription factor 1a |
| chr23_+_30730121 | 9.98 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr10_+_7182423 | 9.98 |
ENSDART00000186788
|
psd3l
|
pleckstrin and Sec7 domain containing 3, like |
| chr17_-_35881841 | 9.85 |
ENSDART00000110040
|
sox11a
|
SRY (sex determining region Y)-box 11a |
| chr13_+_25428677 | 9.82 |
ENSDART00000186284
|
si:dkey-51a16.9
|
si:dkey-51a16.9 |
| chr22_-_20011476 | 9.81 |
ENSDART00000093312
ENSDART00000093310 |
celf5a
|
cugbp, Elav-like family member 5a |
| chr13_-_32906265 | 9.80 |
ENSDART00000113823
ENSDART00000171114 ENSDART00000180035 ENSDART00000137570 |
si:dkey-18j18.3
|
si:dkey-18j18.3 |
| chr10_+_25982212 | 9.77 |
ENSDART00000128292
ENSDART00000108808 |
frem2a
|
Fras1 related extracellular matrix protein 2a |
| chr10_+_7182168 | 9.76 |
ENSDART00000172766
|
psd3l
|
pleckstrin and Sec7 domain containing 3, like |
| chr3_+_52999962 | 9.70 |
ENSDART00000104683
|
pbx4
|
pre-B-cell leukemia transcription factor 4 |
| chr19_+_2275019 | 9.66 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr19_-_10395683 | 9.58 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr22_-_4760187 | 9.57 |
ENSDART00000081969
|
rad23ab
|
RAD23 homolog A, nucleotide excision repair protein b |
| chr5_+_23622177 | 9.54 |
ENSDART00000121504
|
cx27.5
|
connexin 27.5 |
| chr5_+_37504309 | 9.54 |
ENSDART00000165465
|
si:ch1073-224n8.1
|
si:ch1073-224n8.1 |
| chr22_-_3449282 | 9.48 |
ENSDART00000136798
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr16_+_22654481 | 9.41 |
ENSDART00000179762
|
chrnb2b
|
cholinergic receptor, nicotinic, beta 2b |
| chr3_+_34821327 | 9.39 |
ENSDART00000055262
|
cdk5r1a
|
cyclin-dependent kinase 5, regulatory subunit 1a (p35) |
| chr9_-_44295071 | 9.34 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr7_+_56472585 | 9.34 |
ENSDART00000135259
ENSDART00000073596 |
ist1
|
increased sodium tolerance 1 homolog (yeast) |
| chr12_-_2522487 | 9.28 |
ENSDART00000022471
ENSDART00000145213 |
mapk8b
|
mitogen-activated protein kinase 8b |
| chr2_-_42173834 | 9.17 |
ENSDART00000098357
ENSDART00000144707 |
slc39a6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr4_-_12007404 | 9.17 |
ENSDART00000092250
|
btbd11a
|
BTB (POZ) domain containing 11a |
| chr17_+_38295847 | 9.16 |
ENSDART00000008532
|
mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr8_-_30779101 | 9.16 |
ENSDART00000062229
|
p2rx7
|
purinergic receptor P2X, ligand-gated ion channel, 7 |
| chr16_+_17389116 | 9.15 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxp1a+foxp1b+foxp3a_foxg1b_foxp2_foxd1+foxd7_foxo1a+foxo1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.9 | 53.8 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 11.9 | 47.8 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 6.0 | 18.0 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 5.8 | 23.2 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 5.6 | 16.7 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 4.8 | 33.8 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 4.8 | 19.3 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 4.7 | 23.4 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 4.3 | 30.3 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 4.1 | 20.6 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 4.1 | 28.8 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 4.1 | 16.4 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 3.7 | 11.1 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 3.6 | 21.7 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 3.5 | 10.5 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 3.4 | 20.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 3.3 | 16.6 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 3.3 | 29.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 3.2 | 15.9 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 3.0 | 44.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 2.9 | 8.6 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 2.7 | 8.2 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 2.7 | 5.3 | GO:0070528 | protein kinase C signaling(GO:0070528) |
| 2.6 | 15.4 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 2.4 | 36.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 2.4 | 9.5 | GO:1901827 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 2.3 | 31.8 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 2.3 | 6.8 | GO:0000423 | macromitophagy(GO:0000423) |
| 2.2 | 6.7 | GO:0042245 | RNA repair(GO:0042245) |
| 2.2 | 15.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 2.2 | 10.8 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 2.1 | 6.4 | GO:0071514 | genetic imprinting(GO:0071514) |
| 2.0 | 10.2 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 2.0 | 5.9 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 1.9 | 5.7 | GO:1903334 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 1.9 | 43.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 1.9 | 30.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 1.9 | 16.8 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 1.8 | 11.0 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 1.8 | 7.2 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 1.8 | 19.5 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 1.7 | 5.1 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 1.6 | 16.5 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 1.6 | 9.7 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 1.6 | 4.8 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 1.6 | 4.7 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 1.6 | 29.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 1.5 | 9.0 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 1.5 | 22.2 | GO:0001964 | startle response(GO:0001964) |
| 1.4 | 5.7 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 1.4 | 7.0 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 1.4 | 19.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 1.3 | 22.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 1.3 | 4.0 | GO:0006585 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 1.3 | 9.3 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 1.3 | 6.4 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 1.3 | 61.2 | GO:0021854 | hypothalamus development(GO:0021854) |
| 1.2 | 2.5 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 1.2 | 8.4 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 1.2 | 8.3 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 1.2 | 3.5 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 1.2 | 12.9 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 1.2 | 12.8 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 1.2 | 5.8 | GO:0044805 | late nucleophagy(GO:0044805) |
| 1.2 | 5.8 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 1.1 | 5.7 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 1.1 | 3.4 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 1.1 | 39.5 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 1.1 | 4.5 | GO:0035522 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 1.1 | 9.0 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 1.1 | 4.5 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 1.1 | 2.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 1.1 | 4.4 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 1.1 | 11.1 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 1.1 | 28.2 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 1.1 | 3.2 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 1.1 | 4.3 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 1.1 | 6.3 | GO:0030242 | pexophagy(GO:0030242) |
| 1.0 | 5.2 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 1.0 | 18.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 1.0 | 21.5 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 1.0 | 9.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 1.0 | 30.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 1.0 | 8.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 1.0 | 13.0 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 1.0 | 14.7 | GO:0030317 | sperm motility(GO:0030317) |
| 1.0 | 10.6 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 1.0 | 17.3 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.9 | 17.0 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.9 | 10.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.9 | 3.5 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.9 | 25.2 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.9 | 5.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.9 | 3.4 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.8 | 14.4 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.8 | 5.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.8 | 8.4 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.8 | 4.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.8 | 4.1 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.8 | 3.2 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.8 | 3.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.8 | 2.4 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.8 | 6.3 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.8 | 4.7 | GO:0090467 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.8 | 10.1 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.8 | 5.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.8 | 9.2 | GO:0033198 | response to ATP(GO:0033198) |
| 0.7 | 0.7 | GO:0010574 | vascular endothelial growth factor production(GO:0010573) regulation of vascular endothelial growth factor production(GO:0010574) positive regulation of vascular endothelial growth factor production(GO:0010575) |
| 0.7 | 3.7 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.7 | 8.8 | GO:0043279 | response to nicotine(GO:0035094) response to alkaloid(GO:0043279) |
| 0.7 | 2.2 | GO:0009750 | response to fructose(GO:0009750) |
| 0.7 | 21.9 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.7 | 10.2 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.7 | 4.3 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.7 | 7.9 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.7 | 9.5 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.7 | 4.7 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.7 | 2.0 | GO:2000425 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) regulation of apoptotic cell clearance(GO:2000425) |
| 0.7 | 11.9 | GO:0007032 | endosome organization(GO:0007032) |
| 0.7 | 4.6 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.7 | 4.6 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.7 | 16.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.6 | 27.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.6 | 4.3 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.6 | 3.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.6 | 13.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.6 | 3.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.6 | 15.8 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.6 | 4.2 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.6 | 3.6 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.6 | 9.0 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.6 | 7.2 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.6 | 12.4 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.6 | 4.1 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.6 | 7.6 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.6 | 2.3 | GO:0055016 | hypochord development(GO:0055016) |
| 0.6 | 9.9 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.6 | 10.4 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.6 | 6.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.6 | 11.7 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.5 | 2.7 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.5 | 15.3 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.5 | 9.8 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.5 | 18.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.5 | 3.8 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.5 | 9.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.5 | 3.6 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.5 | 8.2 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.5 | 1.5 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.5 | 2.0 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.5 | 1.5 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.5 | 3.5 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.5 | 3.9 | GO:2000780 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.5 | 15.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.5 | 6.8 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.5 | 6.2 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.5 | 1.4 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.5 | 25.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.5 | 19.3 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.5 | 2.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.5 | 3.6 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.4 | 7.2 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.4 | 1.3 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.4 | 2.6 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.4 | 20.6 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.4 | 5.6 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.4 | 0.9 | GO:1904427 | positive regulation of calcium ion transmembrane transport(GO:1904427) |
| 0.4 | 1.7 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.4 | 3.4 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.4 | 6.7 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.4 | 2.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.4 | 4.2 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.4 | 8.4 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.4 | 4.1 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.4 | 6.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.4 | 8.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.4 | 3.7 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.4 | 26.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.4 | 4.4 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.4 | 3.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.4 | 3.9 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.4 | 8.1 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.4 | 5.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.4 | 72.6 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.4 | 12.6 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.4 | 4.9 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.4 | 7.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.4 | 7.8 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.4 | 3.7 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.4 | 10.4 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.4 | 2.2 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.4 | 0.7 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.4 | 5.5 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.4 | 3.7 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.4 | 2.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.4 | 9.7 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.4 | 4.3 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.4 | 3.2 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.4 | 11.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.4 | 2.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) dendritic spine organization(GO:0097061) |
| 0.4 | 1.4 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.3 | 1.0 | GO:0097502 | protein mannosylation(GO:0035268) mannosylation(GO:0097502) |
| 0.3 | 9.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.3 | 6.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 1.7 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.3 | 9.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.3 | 1.0 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.3 | 2.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.3 | 4.9 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.3 | 2.9 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.3 | 18.9 | GO:0007338 | single fertilization(GO:0007338) |
| 0.3 | 12.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.3 | 7.4 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.3 | 8.5 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.3 | 10.2 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.3 | 7.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.3 | 4.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.3 | 1.5 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.3 | 0.9 | GO:1903078 | regulation of Golgi to plasma membrane protein transport(GO:0042996) positive regulation of Golgi to plasma membrane protein transport(GO:0042998) regulation of establishment of protein localization to plasma membrane(GO:0090003) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.3 | 2.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.3 | 4.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.3 | 6.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 3.1 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.3 | 1.4 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.3 | 2.6 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.3 | 1.6 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.3 | 3.9 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.3 | 1.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.3 | 51.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.3 | 1.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 0.2 | GO:0044878 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.2 | 1.0 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.2 | 3.2 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.2 | 1.5 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.2 | 16.4 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.2 | 1.7 | GO:0032387 | negative regulation of intracellular transport(GO:0032387) |
| 0.2 | 0.7 | GO:0072314 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.2 | 1.7 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.2 | 1.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.2 | 2.5 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.2 | 1.8 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.2 | 2.5 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.2 | 1.6 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 2.9 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 4.4 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.2 | 17.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.2 | 17.6 | GO:0042552 | myelination(GO:0042552) |
| 0.2 | 5.2 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.2 | 7.0 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.2 | 0.6 | GO:1904729 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.2 | 5.9 | GO:0030111 | regulation of Wnt signaling pathway(GO:0030111) |
| 0.2 | 20.8 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.2 | 36.8 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.2 | 16.7 | GO:0021782 | glial cell development(GO:0021782) |
| 0.2 | 7.6 | GO:0010906 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.2 | 3.0 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.2 | 0.6 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.2 | 8.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.2 | 1.4 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.2 | 1.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 0.8 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.2 | 4.9 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.2 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 8.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.2 | 10.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 2.0 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 0.5 | GO:0071033 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.2 | 1.0 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.2 | 3.4 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.2 | 5.6 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.2 | 3.5 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.2 | 2.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.2 | 1.0 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.2 | 2.7 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.2 | 6.9 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.2 | 92.6 | GO:0031175 | neuron projection development(GO:0031175) |
| 0.2 | 3.0 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.2 | 2.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.2 | 11.1 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.2 | 1.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 5.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.2 | 4.8 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.2 | 5.9 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.2 | 5.9 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.2 | 0.3 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.2 | 1.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 2.9 | GO:0003140 | determination of left/right asymmetry in lateral mesoderm(GO:0003140) |
| 0.2 | 1.5 | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.1 | 3.6 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.1 | 1.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 14.4 | GO:0030900 | forebrain development(GO:0030900) |
| 0.1 | 6.8 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 1.0 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 12.1 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.1 | 12.0 | GO:0009615 | response to virus(GO:0009615) |
| 0.1 | 0.8 | GO:0015846 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 8.1 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.1 | 1.8 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 7.2 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.1 | 3.8 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 0.3 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.1 | 1.0 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 3.0 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.1 | 6.5 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 1.8 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 0.8 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 1.1 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.1 | 11.6 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.1 | 2.6 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.1 | 2.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 3.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.7 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 10.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 1.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 3.4 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 2.7 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.1 | 0.3 | GO:0006273 | lagging strand elongation(GO:0006273) mitochondrial DNA repair(GO:0043504) |
| 0.1 | 1.5 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.1 | 1.2 | GO:0051984 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of chromosome segregation(GO:0051984) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 2.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.9 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 3.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 2.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 28.8 | GO:0098662 | inorganic cation transmembrane transport(GO:0098662) |
| 0.1 | 0.5 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 3.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 2.5 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.1 | 31.2 | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter(GO:0045944) |
| 0.1 | 1.2 | GO:1903322 | positive regulation of protein ubiquitination(GO:0031398) positive regulation of protein modification by small protein conjugation or removal(GO:1903322) |
| 0.1 | 4.1 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 18.0 | GO:0006897 | endocytosis(GO:0006897) |
| 0.1 | 7.1 | GO:1990778 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.1 | 0.6 | GO:0043535 | regulation of endothelial cell migration(GO:0010594) regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.1 | 0.9 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 6.5 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.1 | 4.2 | GO:0008037 | cell recognition(GO:0008037) |
| 0.1 | 3.5 | GO:0030509 | BMP signaling pathway(GO:0030509) response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
| 0.1 | 3.8 | GO:0006821 | chloride transport(GO:0006821) |
| 0.1 | 23.8 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.1 | 1.1 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 2.1 | GO:0098840 | intraciliary transport(GO:0042073) protein transport along microtubule(GO:0098840) |
| 0.1 | 0.7 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 2.7 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.1 | 3.0 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.1 | 2.8 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 0.8 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 1.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 12.8 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.1 | 1.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 4.0 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.1 | 7.5 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
| 0.1 | 0.8 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 1.3 | GO:0035282 | somitogenesis(GO:0001756) segmentation(GO:0035282) |
| 0.1 | 1.4 | GO:0035966 | response to topologically incorrect protein(GO:0035966) |
| 0.1 | 4.7 | GO:0060070 | canonical Wnt signaling pathway(GO:0060070) |
| 0.0 | 1.0 | GO:0045664 | regulation of neuron differentiation(GO:0045664) |
| 0.0 | 0.2 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 2.2 | GO:0033334 | fin morphogenesis(GO:0033334) |
| 0.0 | 1.3 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.0 | 1.5 | GO:0042471 | ear morphogenesis(GO:0042471) inner ear morphogenesis(GO:0042472) |
| 0.0 | 2.1 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 1.9 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 1.3 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 0.3 | GO:0007096 | regulation of exit from mitosis(GO:0007096) exit from mitosis(GO:0010458) |
| 0.0 | 1.4 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 1.5 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 1.4 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 2.0 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 1.0 | GO:0030301 | sterol transport(GO:0015918) cholesterol transport(GO:0030301) |
| 0.0 | 0.2 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 1.0 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.0 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 1.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 5.5 | GO:0007420 | brain development(GO:0007420) |
| 0.0 | 0.4 | GO:0006282 | regulation of DNA repair(GO:0006282) |
| 0.0 | 1.5 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.0 | 1.8 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.8 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.1 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.0 | 3.0 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 2.4 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.7 | GO:0019362 | pyridine nucleotide metabolic process(GO:0019362) nicotinamide nucleotide metabolic process(GO:0046496) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.7 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:0006826 | iron ion transport(GO:0006826) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 73.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 6.0 | 36.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 4.9 | 24.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 4.1 | 16.4 | GO:0097268 | cytoophidium(GO:0097268) |
| 3.8 | 11.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 3.3 | 3.3 | GO:0044297 | cell body(GO:0044297) |
| 3.0 | 26.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 3.0 | 20.7 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 2.9 | 14.6 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 2.9 | 14.4 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 2.6 | 12.8 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 2.4 | 24.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 2.1 | 19.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 2.1 | 71.5 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 2.1 | 32.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 1.9 | 5.7 | GO:0072380 | TRC complex(GO:0072380) |
| 1.8 | 5.5 | GO:0031213 | RSF complex(GO:0031213) |
| 1.7 | 17.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 1.7 | 6.8 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 1.6 | 6.6 | GO:0016460 | myosin II complex(GO:0016460) |
| 1.6 | 22.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 1.6 | 7.9 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 1.5 | 7.5 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 1.4 | 5.6 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 1.3 | 5.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 1.3 | 54.8 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 1.2 | 38.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 1.2 | 25.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 1.0 | 16.6 | GO:0030315 | T-tubule(GO:0030315) |
| 1.0 | 22.0 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 1.0 | 31.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 1.0 | 33.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.9 | 5.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.9 | 3.7 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.9 | 14.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.9 | 7.4 | GO:0071914 | prominosome(GO:0071914) |
| 0.9 | 29.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.8 | 3.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.8 | 5.6 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.8 | 1.6 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.7 | 3.0 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.7 | 5.8 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.7 | 6.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.7 | 9.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.7 | 53.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.7 | 3.9 | GO:0000801 | central element(GO:0000801) |
| 0.7 | 5.2 | GO:0016586 | RSC complex(GO:0016586) |
| 0.6 | 11.7 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.6 | 11.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.6 | 26.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.6 | 3.0 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.6 | 1.2 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.6 | 3.4 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.5 | 7.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.5 | 4.8 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.5 | 6.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.5 | 12.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.5 | 4.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.5 | 2.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.5 | 7.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.5 | 29.8 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.5 | 4.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.5 | 15.2 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.5 | 2.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.5 | 3.7 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.4 | 80.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.4 | 10.2 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.4 | 3.5 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.4 | 48.6 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.4 | 6.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.4 | 2.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.4 | 5.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.4 | 8.1 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.4 | 11.6 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.4 | 2.5 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.4 | 24.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.4 | 1.4 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.3 | 8.1 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.3 | 2.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.3 | 2.2 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.3 | 2.4 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.3 | 15.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 7.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.3 | 6.4 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.3 | 5.8 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.3 | 10.2 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.3 | 27.5 | GO:0070160 | bicellular tight junction(GO:0005923) apical junction complex(GO:0043296) occluding junction(GO:0070160) |
| 0.3 | 26.2 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.3 | 1.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 19.4 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.2 | 68.2 | GO:0005912 | adherens junction(GO:0005912) |
| 0.2 | 22.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 3.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 11.7 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.2 | 3.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 3.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 28.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.2 | 2.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 12.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 20.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 5.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 2.3 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.2 | 2.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 0.8 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 5.8 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.2 | 1.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 7.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.2 | 2.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 1.4 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.2 | 9.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 1.0 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 1.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 12.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 2.3 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.2 | 77.6 | GO:0043005 | neuron projection(GO:0043005) |
| 0.1 | 1.2 | GO:0030670 | endocytic vesicle membrane(GO:0030666) phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 2.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 10.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 8.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 7.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 5.5 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 1.7 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 4.4 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 1.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 12.8 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 0.5 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 2.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 15.1 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 1.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.8 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 32.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 1.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.5 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 9.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 121.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 1.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 2.6 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 34.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 7.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 52.6 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.1 | 4.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.5 | GO:0060091 | kinocilium(GO:0060091) kinociliary basal body(GO:1902636) |
| 0.1 | 1.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.5 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 1.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 13.2 | GO:0042995 | cell projection(GO:0042995) |
| 0.1 | 0.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 2.1 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 3.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 4.4 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 59.9 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.0 | 1.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.0 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.4 | GO:0016459 | myosin complex(GO:0016459) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.9 | 47.8 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 7.7 | 23.2 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 6.0 | 36.0 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 5.7 | 45.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 4.1 | 16.4 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 4.0 | 24.2 | GO:0097363 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 3.8 | 26.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 3.6 | 14.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 3.4 | 20.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 3.2 | 16.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 2.9 | 14.4 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 2.8 | 11.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 2.7 | 64.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 2.5 | 35.4 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 2.5 | 7.5 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 2.4 | 7.2 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 2.4 | 9.5 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 2.2 | 37.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 2.2 | 15.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 2.2 | 11.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 2.1 | 33.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 2.1 | 10.4 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 2.0 | 35.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 1.9 | 24.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.8 | 36.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 1.8 | 14.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.7 | 8.6 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 1.7 | 10.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 1.7 | 6.7 | GO:0043734 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 1.7 | 39.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 1.6 | 6.6 | GO:0032034 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 1.6 | 53.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 1.6 | 7.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 1.6 | 23.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 1.5 | 32.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 1.5 | 26.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 1.5 | 29.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 1.4 | 7.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 1.4 | 14.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.4 | 5.6 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 1.4 | 12.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.4 | 17.9 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 1.3 | 40.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 1.3 | 9.3 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 1.3 | 18.5 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 1.3 | 4.0 | GO:0052855 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 1.3 | 5.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 1.3 | 10.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 1.3 | 11.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.2 | 32.6 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 1.2 | 18.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 1.1 | 7.9 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 1.1 | 15.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 1.1 | 8.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.1 | 5.4 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 1.1 | 4.3 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 1.0 | 8.2 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 1.0 | 5.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 1.0 | 11.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 1.0 | 3.9 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.9 | 20.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.9 | 5.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.9 | 3.7 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.9 | 16.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.9 | 4.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.9 | 3.5 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.9 | 4.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.8 | 5.9 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.8 | 10.8 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.8 | 4.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.8 | 4.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.8 | 3.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.8 | 15.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.8 | 3.1 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.8 | 9.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.8 | 2.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.8 | 7.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.7 | 19.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.7 | 13.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.7 | 6.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.7 | 4.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.7 | 14.0 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.7 | 6.9 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.7 | 8.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.7 | 4.7 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.7 | 34.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.7 | 4.7 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.7 | 2.6 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.6 | 3.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.6 | 16.3 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.6 | 16.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.6 | 12.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.6 | 4.9 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.6 | 1.8 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.6 | 9.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.6 | 4.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.6 | 5.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.6 | 23.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.6 | 2.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.6 | 1.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.6 | 5.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.5 | 1.6 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.5 | 5.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.5 | 21.7 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.5 | 1.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.5 | 25.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.5 | 2.7 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.5 | 3.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.5 | 26.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.5 | 1.6 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.5 | 3.7 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.5 | 12.6 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.5 | 6.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.5 | 4.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.5 | 7.2 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.5 | 2.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.5 | 4.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.5 | 19.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.5 | 54.9 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.5 | 15.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.4 | 4.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.4 | 4.0 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.4 | 2.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.4 | 3.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.4 | 3.3 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.4 | 15.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.4 | 7.8 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.4 | 10.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.4 | 2.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.4 | 10.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.3 | 8.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.3 | 2.0 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.3 | 1.7 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.3 | 1.7 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.3 | 4.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 4.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 2.4 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.3 | 8.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 0.9 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.3 | 0.9 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 3.0 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.3 | 2.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 2.0 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.3 | 5.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.3 | 11.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.3 | 6.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.3 | 6.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.3 | 1.3 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.3 | 3.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 2.9 | GO:0030955 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.3 | 4.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.3 | 2.6 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.3 | 91.8 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.2 | 3.5 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 41.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.2 | 1.5 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 5.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 10.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 4.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.2 | 1.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 2.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 20.5 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.2 | 28.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.2 | 39.9 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.2 | 26.4 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.2 | 4.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 11.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.2 | 1.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 3.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 3.0 | GO:0016208 | AMP binding(GO:0016208) |
| 0.2 | 1.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 5.3 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.2 | 0.7 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.2 | 0.9 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.2 | 0.5 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.2 | 8.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 0.7 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 4.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.2 | 1.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 1.8 | GO:0032182 | ubiquitin-like protein binding(GO:0032182) |
| 0.2 | 4.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 1.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 7.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 32.3 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.1 | 18.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.8 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 21.0 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 5.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 2.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 1.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 2.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 2.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 2.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 42.6 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 4.1 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.1 | 4.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 3.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 4.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.0 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 0.4 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.1 | 41.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 3.0 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 3.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.8 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 2.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.6 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.3 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.8 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 1.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 4.7 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.1 | 4.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.3 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 1.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 3.9 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 0.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 3.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 2.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.5 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 1.5 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.1 | 0.2 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 13.1 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.1 | 4.8 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.1 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 2.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 1.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.9 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.6 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 1.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 2.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 1.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 1.0 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 8.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 35.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 1.2 | 25.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 1.1 | 9.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 1.0 | 43.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 1.0 | 22.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 1.0 | 9.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.9 | 52.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.8 | 11.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.7 | 6.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.7 | 14.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.5 | 8.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.4 | 5.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.4 | 5.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.4 | 21.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.3 | 1.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.3 | 9.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.3 | 4.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.3 | 3.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.3 | 9.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 7.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 2.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 4.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.2 | 4.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.2 | 3.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 1.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.2 | 3.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 2.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 4.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 2.3 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 0.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 1.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 44.6 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 4.1 | 20.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 4.1 | 61.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 3.5 | 59.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 1.8 | 9.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.6 | 34.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 1.2 | 14.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 1.1 | 54.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.9 | 8.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.8 | 10.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.7 | 8.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.7 | 7.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.7 | 4.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.6 | 11.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.6 | 5.7 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.6 | 4.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.6 | 7.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.6 | 8.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.6 | 5.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.5 | 5.9 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.5 | 5.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.4 | 4.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.4 | 5.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.4 | 3.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.4 | 6.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.4 | 5.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.4 | 12.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.3 | 5.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 2.2 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.3 | 3.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.3 | 3.7 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.3 | 2.9 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.3 | 4.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 2.5 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.3 | 12.8 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.3 | 1.3 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.3 | 2.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 13.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 0.7 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.2 | 2.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 7.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 1.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 3.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 5.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 2.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 6.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 2.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 3.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 4.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 1.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 3.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.3 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 12.5 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 2.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 1.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.7 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 4.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.0 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 0.9 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 0.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 5.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 1.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |