Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
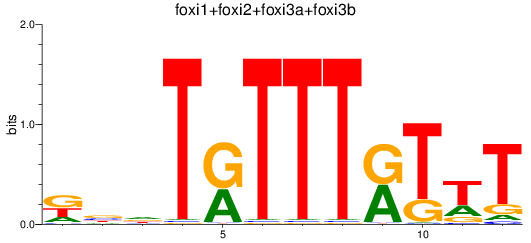
Results for foxi1+foxi2+foxi3a+foxi3b
Z-value: 1.31
Transcription factors associated with foxi1+foxi2+foxi3a+foxi3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxi3b
|
ENSDARG00000009550 | forkhead box I3b |
|
foxi3a
|
ENSDARG00000055926 | forkhead box I3a |
|
foxi2
|
ENSDARG00000069715 | forkhead box I2 |
|
foxi1
|
ENSDARG00000104566 | forkhead box i1 |
|
foxi3b
|
ENSDARG00000112022 | forkhead box I3b |
|
foxi3b
|
ENSDARG00000112303 | forkhead box I3b |
|
foxi2
|
ENSDARG00000116791 | forkhead box I2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxi3a | dr11_v1_chr21_+_30351256_30351256 | -0.50 | 2.2e-07 | Click! |
| foxi3b | dr11_v1_chr14_+_34514336_34514336 | -0.47 | 1.9e-06 | Click! |
| foxi1 | dr11_v1_chr12_-_43664682_43664682 | 0.35 | 6.0e-04 | Click! |
| foxi2 | dr11_v1_chr13_+_13578552_13578552 | -0.20 | 5.9e-02 | Click! |
Activity profile of foxi1+foxi2+foxi3a+foxi3b motif
Sorted Z-values of foxi1+foxi2+foxi3a+foxi3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_37837245 | 34.17 |
ENSDART00000171617
|
epd
|
ependymin |
| chr6_-_35446110 | 22.30 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr14_-_32016615 | 19.74 |
ENSDART00000105761
|
zic3
|
zic family member 3 heterotaxy 1 (odd-paired homolog, Drosophila) |
| chr4_+_8797197 | 18.75 |
ENSDART00000158671
|
sult4a1
|
sulfotransferase family 4A, member 1 |
| chr8_-_14049404 | 17.49 |
ENSDART00000093117
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr16_-_17207754 | 16.31 |
ENSDART00000063804
|
wu:fj39g12
|
wu:fj39g12 |
| chr10_-_24343507 | 15.77 |
ENSDART00000002974
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr10_+_17850934 | 14.65 |
ENSDART00000113666
ENSDART00000145936 |
phf24
|
PHD finger protein 24 |
| chr24_-_17444067 | 14.51 |
ENSDART00000155843
|
cntnap2a
|
contactin associated protein like 2a |
| chr5_+_36768674 | 13.90 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr13_+_27316934 | 13.62 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr7_-_52842605 | 13.18 |
ENSDART00000083002
|
map1aa
|
microtubule-associated protein 1Aa |
| chr12_+_5081759 | 13.09 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr5_-_33215261 | 11.90 |
ENSDART00000097935
ENSDART00000134777 |
si:dkey-226m8.10
|
si:dkey-226m8.10 |
| chr20_+_30445971 | 11.74 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr12_+_13405445 | 11.56 |
ENSDART00000089042
|
kcnh4b
|
potassium voltage-gated channel, subfamily H (eag-related), member 4b |
| chr11_-_17713987 | 11.49 |
ENSDART00000090401
|
fam19a4b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4b |
| chr21_-_42202792 | 11.19 |
ENSDART00000124708
|
gabra6b
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6b |
| chr14_-_2352384 | 10.79 |
ENSDART00000170666
|
si:ch73-233f7.7
|
si:ch73-233f7.7 |
| chr6_-_10780698 | 10.72 |
ENSDART00000151714
|
gpr155b
|
G protein-coupled receptor 155b |
| chr14_-_32403554 | 10.69 |
ENSDART00000172873
ENSDART00000173408 ENSDART00000173114 ENSDART00000185594 ENSDART00000186762 ENSDART00000010982 |
fgf13a
|
fibroblast growth factor 13a |
| chr17_+_15534815 | 10.35 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr9_-_33785093 | 10.16 |
ENSDART00000140779
ENSDART00000059837 |
fundc1
|
FUN14 domain containing 1 |
| chr25_-_11088839 | 9.98 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr10_+_21730585 | 9.88 |
ENSDART00000188576
|
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr11_+_21586335 | 9.79 |
ENSDART00000091182
|
FAM72B
|
zgc:101564 |
| chr16_-_29334672 | 9.75 |
ENSDART00000162835
|
bcan
|
brevican |
| chr16_+_22654481 | 9.65 |
ENSDART00000179762
|
chrnb2b
|
cholinergic receptor, nicotinic, beta 2b |
| chr1_-_30039331 | 9.64 |
ENSDART00000086935
ENSDART00000143800 |
MARCH4 (1 of many)
|
zgc:153256 |
| chr1_-_22757145 | 9.64 |
ENSDART00000134719
|
prom1b
|
prominin 1 b |
| chr5_-_61942482 | 9.48 |
ENSDART00000097338
|
napaa
|
N-ethylmaleimide-sensitive factor attachment protein, alpha a |
| chr20_-_26042070 | 9.46 |
ENSDART00000140255
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr6_+_39222598 | 9.39 |
ENSDART00000154991
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr14_-_18671334 | 9.10 |
ENSDART00000182381
|
slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr3_-_43356082 | 8.89 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr19_-_44091405 | 8.78 |
ENSDART00000132800
|
rad21b
|
RAD21 cohesin complex component b |
| chr12_+_13404784 | 8.61 |
ENSDART00000167977
|
kcnh4b
|
potassium voltage-gated channel, subfamily H (eag-related), member 4b |
| chr2_+_5563077 | 8.51 |
ENSDART00000111220
|
mb21d2
|
Mab-21 domain containing 2 |
| chr25_-_14087377 | 8.45 |
ENSDART00000124140
|
zgc:101566
|
zgc:101566 |
| chr5_-_23362602 | 8.23 |
ENSDART00000137120
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr14_-_21219659 | 7.80 |
ENSDART00000089867
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr14_-_2348917 | 7.79 |
ENSDART00000159004
|
si:ch73-233f7.8
|
si:ch73-233f7.8 |
| chr19_-_27261102 | 7.54 |
ENSDART00000143919
|
gabbr1b
|
gamma-aminobutyric acid (GABA) B receptor, 1b |
| chr2_-_37043905 | 7.27 |
ENSDART00000056514
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr2_-_37043540 | 7.25 |
ENSDART00000131834
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr16_-_563235 | 7.20 |
ENSDART00000016303
|
irx2a
|
iroquois homeobox 2a |
| chr19_-_9503473 | 7.07 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr10_+_1849874 | 7.07 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr14_+_34966598 | 7.05 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr11_+_30817943 | 6.86 |
ENSDART00000150130
ENSDART00000159997 |
cacna1ab
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, b |
| chr6_-_39167732 | 6.79 |
ENSDART00000153626
|
apof
|
apolipoprotein F |
| chr12_+_26467847 | 6.77 |
ENSDART00000022495
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr21_-_33995213 | 6.69 |
ENSDART00000140184
|
EBF1 (1 of many)
|
si:ch211-51e8.2 |
| chr22_+_31821815 | 6.66 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr10_+_1638876 | 6.64 |
ENSDART00000184484
ENSDART00000060946 ENSDART00000181251 |
sgsm1b
|
small G protein signaling modulator 1b |
| chr7_-_49801183 | 6.51 |
ENSDART00000052083
|
fjx1
|
four jointed box 1 |
| chr6_-_6448519 | 6.46 |
ENSDART00000180157
ENSDART00000191112 |
si:ch211-194e18.2
|
si:ch211-194e18.2 |
| chr21_+_31434251 | 6.44 |
ENSDART00000040740
ENSDART00000130157 |
si:ch211-12m10.1
si:ch211-166i24.1
|
si:ch211-12m10.1 si:ch211-166i24.1 |
| chr10_+_21584195 | 6.27 |
ENSDART00000100599
|
pcdh1a3
|
protocadherin 1 alpha 3 |
| chr13_+_23157053 | 6.21 |
ENSDART00000162359
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr19_+_2275019 | 6.21 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr13_-_32626247 | 6.09 |
ENSDART00000100663
|
wdr35
|
WD repeat domain 35 |
| chr19_-_8880688 | 6.06 |
ENSDART00000039629
|
celf3a
|
cugbp, Elav-like family member 3a |
| chr1_+_6817292 | 6.06 |
ENSDART00000145822
ENSDART00000092118 |
erbb4a
|
erb-b2 receptor tyrosine kinase 4a |
| chr1_-_50611031 | 6.03 |
ENSDART00000148285
|
ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr18_-_20560007 | 6.02 |
ENSDART00000141367
ENSDART00000090186 |
si:ch211-238n5.4
|
si:ch211-238n5.4 |
| chr2_-_12243213 | 6.01 |
ENSDART00000113081
|
gpr158b
|
G protein-coupled receptor 158b |
| chr7_+_52766211 | 5.99 |
ENSDART00000186191
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr24_-_9002038 | 5.96 |
ENSDART00000066783
ENSDART00000150185 |
mppe1
|
metallophosphoesterase 1 |
| chr2_-_31798717 | 5.83 |
ENSDART00000170880
|
retreg1
|
reticulophagy regulator 1 |
| chr5_+_49744713 | 5.70 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr21_-_33995710 | 5.69 |
ENSDART00000100508
ENSDART00000179622 |
ebf1b
|
early B cell factor 1b |
| chr5_+_42912966 | 5.57 |
ENSDART00000039973
|
rufy3
|
RUN and FYVE domain containing 3 |
| chr2_+_3595333 | 5.49 |
ENSDART00000041052
|
c1ql3b
|
complement component 1, q subcomponent-like 3b |
| chr15_-_18432673 | 5.43 |
ENSDART00000146853
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr2_+_31437547 | 5.29 |
ENSDART00000141170
|
stam
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr4_-_12978925 | 5.24 |
ENSDART00000013839
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr18_-_26126258 | 5.19 |
ENSDART00000145875
|
ANKRD34C
|
ankyrin repeat domain 34C |
| chr8_+_23147218 | 5.13 |
ENSDART00000030920
ENSDART00000141175 ENSDART00000146264 |
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr7_+_24023653 | 5.12 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr15_-_20024205 | 5.12 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr15_+_28940352 | 5.10 |
ENSDART00000154085
|
gipr
|
gastric inhibitory polypeptide receptor |
| chr14_-_21218891 | 4.98 |
ENSDART00000158294
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr11_-_21586157 | 4.83 |
ENSDART00000190095
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr7_-_49800755 | 4.83 |
ENSDART00000180072
|
fjx1
|
four jointed box 1 |
| chr6_+_46431848 | 4.76 |
ENSDART00000181056
ENSDART00000144569 ENSDART00000064865 ENSDART00000133992 |
stau1
|
staufen double-stranded RNA binding protein 1 |
| chr1_+_2190714 | 4.63 |
ENSDART00000132126
|
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr22_-_14367966 | 4.61 |
ENSDART00000188796
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr7_-_33868903 | 4.56 |
ENSDART00000173500
ENSDART00000178746 |
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr12_-_35505610 | 4.47 |
ENSDART00000105518
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr19_+_20724347 | 4.43 |
ENSDART00000090757
|
kat2b
|
K(lysine) acetyltransferase 2B |
| chr5_-_48070779 | 4.42 |
ENSDART00000078401
|
tmem161b
|
transmembrane protein 161B |
| chr6_-_12270226 | 4.41 |
ENSDART00000180473
|
pkp4
|
plakophilin 4 |
| chr10_-_43404027 | 4.37 |
ENSDART00000086227
|
edil3b
|
EGF-like repeats and discoidin I-like domains 3b |
| chr15_+_23657051 | 4.19 |
ENSDART00000078336
|
klc3
|
kinesin light chain 3 |
| chr23_+_21479958 | 4.17 |
ENSDART00000188302
ENSDART00000144320 |
si:dkey-1c11.1
|
si:dkey-1c11.1 |
| chr20_-_31252809 | 4.10 |
ENSDART00000137236
|
hpcal1
|
hippocalcin-like 1 |
| chr7_+_7511914 | 4.06 |
ENSDART00000172848
|
clcn3
|
chloride channel 3 |
| chr19_-_27462720 | 4.05 |
ENSDART00000135901
|
si:ch73-25f10.6
|
si:ch73-25f10.6 |
| chr15_-_33807758 | 4.02 |
ENSDART00000158445
|
pds5b
|
PDS5 cohesin associated factor B |
| chr21_-_21148623 | 4.00 |
ENSDART00000184364
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr7_-_52334429 | 3.96 |
ENSDART00000187372
|
CR938716.1
|
|
| chr10_+_19017146 | 3.90 |
ENSDART00000038674
|
tmem230a
|
transmembrane protein 230a |
| chr14_+_7932973 | 3.85 |
ENSDART00000109941
|
cxxc5b
|
CXXC finger protein 5b |
| chr24_-_18659147 | 3.81 |
ENSDART00000166039
|
zgc:66014
|
zgc:66014 |
| chr9_-_38398789 | 3.79 |
ENSDART00000188384
|
znf142
|
zinc finger protein 142 |
| chr12_-_14211067 | 3.73 |
ENSDART00000077903
|
avl9
|
AVL9 homolog (S. cerevisiase) |
| chr7_+_52712807 | 3.72 |
ENSDART00000174095
ENSDART00000174377 ENSDART00000174061 ENSDART00000174094 ENSDART00000110906 ENSDART00000174071 ENSDART00000174238 |
znf280d
|
zinc finger protein 280D |
| chr23_-_31372639 | 3.71 |
ENSDART00000179908
ENSDART00000135620 ENSDART00000053367 |
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr16_+_5612547 | 3.69 |
ENSDART00000140226
ENSDART00000189352 |
CYTH2
|
si:dkey-283b15.4 |
| chr1_+_33647682 | 3.62 |
ENSDART00000114384
|
znf654
|
zinc finger protein 654 |
| chr19_-_12965020 | 3.60 |
ENSDART00000128975
|
slc25a32a
|
solute carrier family 25 (mitochondrial folate carrier), member 32a |
| chr5_-_37935607 | 3.58 |
ENSDART00000141233
ENSDART00000084868 ENSDART00000125857 |
scn4bb
|
sodium channel, voltage-gated, type IV, beta b |
| chr23_-_42810664 | 3.54 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr14_+_28438947 | 3.52 |
ENSDART00000006489
|
acsl4a
|
acyl-CoA synthetase long chain family member 4a |
| chr3_+_22578369 | 3.52 |
ENSDART00000187695
ENSDART00000182678 ENSDART00000112270 |
tanc2a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2a |
| chr5_+_61361815 | 3.32 |
ENSDART00000009507
|
gatsl2
|
GATS protein-like 2 |
| chr1_-_46862190 | 3.29 |
ENSDART00000145167
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr5_+_51079504 | 3.29 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr4_-_9609634 | 3.25 |
ENSDART00000067188
|
dclre1c
|
DNA cross-link repair 1C, PSO2 homolog (S. cerevisiae) |
| chr18_-_7137153 | 3.23 |
ENSDART00000019571
|
cd9a
|
CD9 molecule a |
| chr10_-_41940302 | 3.22 |
ENSDART00000033121
|
morn3
|
MORN repeat containing 3 |
| chr18_+_27077853 | 3.20 |
ENSDART00000125326
ENSDART00000192660 ENSDART00000098334 |
ppp1r15b
|
protein phosphatase 1, regulatory subunit 15B |
| chr9_+_41040294 | 3.17 |
ENSDART00000000693
|
osgepl1
|
O-sialoglycoprotein endopeptidase-like 1 |
| chr12_-_13966184 | 3.15 |
ENSDART00000066368
|
klhl11
|
kelch-like family member 11 |
| chr5_+_60919378 | 3.14 |
ENSDART00000184915
|
doc2b
|
double C2-like domains, beta |
| chr16_+_28932038 | 3.09 |
ENSDART00000149480
|
npr1b
|
natriuretic peptide receptor 1b |
| chr12_+_18544159 | 3.07 |
ENSDART00000162900
|
mlst8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr23_-_24682244 | 3.04 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr12_-_31422433 | 2.95 |
ENSDART00000186075
ENSDART00000153172 ENSDART00000066256 |
vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr2_+_47471943 | 2.91 |
ENSDART00000141974
|
acsl3b
|
acyl-CoA synthetase long chain family member 3b |
| chr8_+_13368150 | 2.88 |
ENSDART00000114699
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr19_+_1510971 | 2.87 |
ENSDART00000157721
|
SLC45A4 (1 of many)
|
solute carrier family 45 member 4 |
| chr8_-_16738204 | 2.86 |
ENSDART00000100706
|
ndufaf2
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 2 |
| chr5_-_19400166 | 2.84 |
ENSDART00000008994
|
foxn4
|
forkhead box N4 |
| chr18_+_16192083 | 2.77 |
ENSDART00000133042
|
lrriq1
|
leucine-rich repeats and IQ motif containing 1 |
| chr25_+_15647993 | 2.77 |
ENSDART00000186578
ENSDART00000031828 |
spon1b
|
spondin 1b |
| chr13_-_5568928 | 2.76 |
ENSDART00000192679
|
meis1b
|
Meis homeobox 1 b |
| chr14_+_49382180 | 2.73 |
ENSDART00000158329
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr2_+_47471647 | 2.71 |
ENSDART00000184199
|
acsl3b
|
acyl-CoA synthetase long chain family member 3b |
| chr1_+_45839927 | 2.71 |
ENSDART00000148086
ENSDART00000180413 ENSDART00000048191 ENSDART00000179047 |
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr4_+_6833583 | 2.70 |
ENSDART00000165179
ENSDART00000186134 ENSDART00000174507 |
dock4b
|
dedicator of cytokinesis 4b |
| chr18_-_21725638 | 2.70 |
ENSDART00000089853
|
fan1
|
FANCD2/FANCI-associated nuclease 1 |
| chr16_-_5612480 | 2.57 |
ENSDART00000157944
|
CR848788.1
|
|
| chr6_-_14038804 | 2.53 |
ENSDART00000184606
ENSDART00000184609 |
etv5b
|
ets variant 5b |
| chr20_-_4883673 | 2.50 |
ENSDART00000145540
ENSDART00000053877 |
zdhhc14
|
zinc finger, DHHC-type containing 14 |
| chr10_-_11840353 | 2.46 |
ENSDART00000127581
|
trim23
|
tripartite motif containing 23 |
| chr15_+_21712328 | 2.44 |
ENSDART00000192553
|
NKAPD1
|
zgc:162339 |
| chr22_-_21755524 | 2.43 |
ENSDART00000149635
|
tle2b
|
transducin like enhancer of split 2b |
| chr8_-_7567815 | 2.43 |
ENSDART00000132536
|
hcfc1b
|
host cell factor C1b |
| chr7_+_29080684 | 2.42 |
ENSDART00000173709
ENSDART00000173576 |
acd
|
ACD, shelterin complex subunit and telomerase recruitment factor |
| chr12_-_43428542 | 2.41 |
ENSDART00000192266
|
ptprea
|
protein tyrosine phosphatase, receptor type, E, a |
| chr22_-_28777557 | 2.41 |
ENSDART00000135214
ENSDART00000131761 ENSDART00000005112 |
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr16_-_24561354 | 2.32 |
ENSDART00000193278
ENSDART00000126274 |
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr22_+_10752787 | 2.29 |
ENSDART00000186542
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr20_-_45062514 | 2.29 |
ENSDART00000183529
ENSDART00000182955 |
klhl29
|
kelch-like family member 29 |
| chr10_+_26972755 | 2.22 |
ENSDART00000042162
|
tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr15_+_42431198 | 2.19 |
ENSDART00000189951
ENSDART00000089694 |
tiam1b
|
T cell lymphoma invasion and metastasis 1b |
| chr7_+_42206847 | 2.19 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr9_-_7238839 | 2.17 |
ENSDART00000142726
|
creg2
|
cellular repressor of E1A-stimulated genes 2 |
| chr19_+_16064439 | 2.17 |
ENSDART00000151169
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr6_-_40922971 | 2.16 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr13_-_30645965 | 2.14 |
ENSDART00000109307
|
zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr10_+_34315719 | 2.14 |
ENSDART00000135303
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr9_-_38399432 | 2.13 |
ENSDART00000148268
|
znf142
|
zinc finger protein 142 |
| chr14_-_6943934 | 2.12 |
ENSDART00000126279
|
clk4a
|
CDC-like kinase 4a |
| chr23_-_27822920 | 2.10 |
ENSDART00000023094
|
acvr1ba
|
activin A receptor type 1Ba |
| chr22_-_28777374 | 2.10 |
ENSDART00000188206
|
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr12_+_34854562 | 2.08 |
ENSDART00000130366
|
si:dkey-21c1.4
|
si:dkey-21c1.4 |
| chr19_+_29303847 | 2.08 |
ENSDART00000009149
|
maco1a
|
macoilin 1a |
| chr16_-_13622794 | 2.07 |
ENSDART00000146953
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr8_-_18316482 | 2.06 |
ENSDART00000190156
|
rnf220b
|
ring finger protein 220b |
| chr5_+_24063046 | 2.03 |
ENSDART00000051548
ENSDART00000133355 ENSDART00000142268 |
gps2
|
G protein pathway suppressor 2 |
| chr23_+_33718602 | 1.93 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr17_-_38778826 | 1.91 |
ENSDART00000168182
ENSDART00000124041 ENSDART00000136921 |
dglucy
|
D-glutamate cyclase |
| chr5_-_56924747 | 1.88 |
ENSDART00000014028
|
ppm1db
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Db |
| chr13_+_4368504 | 1.85 |
ENSDART00000108935
|
sft2d1
|
SFT2 domain containing 1 |
| chr4_-_18436899 | 1.85 |
ENSDART00000141671
|
socs2
|
suppressor of cytokine signaling 2 |
| chr7_+_49800849 | 1.82 |
ENSDART00000164838
ENSDART00000187888 |
PAMR1 (1 of many)
|
si:ch211-102l7.3 |
| chr2_-_24962002 | 1.81 |
ENSDART00000132050
|
hltf
|
helicase-like transcription factor |
| chr5_-_49951106 | 1.79 |
ENSDART00000135954
|
fam172a
|
family with sequence similarity 172, member A |
| chr17_+_25856671 | 1.78 |
ENSDART00000064817
|
wapla
|
WAPL cohesin release factor a |
| chr10_+_26973063 | 1.75 |
ENSDART00000143162
ENSDART00000186210 |
tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr15_+_3674765 | 1.75 |
ENSDART00000081802
|
NEK3
|
NIMA related kinase 3 |
| chr7_+_38808027 | 1.70 |
ENSDART00000052323
|
harbi1
|
harbinger transposase derived 1 |
| chr17_-_14966384 | 1.66 |
ENSDART00000105064
|
txndc16
|
thioredoxin domain containing 16 |
| chr16_+_29558530 | 1.65 |
ENSDART00000088146
|
ensab
|
endosulfine alpha b |
| chr4_-_14926637 | 1.61 |
ENSDART00000110199
|
prdm4
|
PR domain containing 4 |
| chr10_-_31563049 | 1.59 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr22_+_17499905 | 1.59 |
ENSDART00000061366
ENSDART00000133793 |
rab11ba
|
RAB11B, member RAS oncogene family, a |
| chr16_+_18535618 | 1.59 |
ENSDART00000021596
|
rxrbb
|
retinoid x receptor, beta b |
| chr9_-_27719998 | 1.58 |
ENSDART00000161068
ENSDART00000148195 ENSDART00000138386 |
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr23_-_33680265 | 1.56 |
ENSDART00000138416
|
tfcp2
|
transcription factor CP2 |
| chr18_-_5111449 | 1.54 |
ENSDART00000046902
|
pdcd10a
|
programmed cell death 10a |
| chr22_+_10752511 | 1.46 |
ENSDART00000081188
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr22_-_35063526 | 1.45 |
ENSDART00000162211
|
zgc:63733
|
zgc:63733 |
| chr14_-_33177935 | 1.45 |
ENSDART00000180583
ENSDART00000078856 |
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr11_-_36350421 | 1.45 |
ENSDART00000141477
|
psma5
|
proteasome subunit alpha 5 |
| chr13_-_29505946 | 1.41 |
ENSDART00000026679
|
cdhr1a
|
cadherin-related family member 1a |
| chr22_-_20720427 | 1.39 |
ENSDART00000105532
|
oaz1a
|
ornithine decarboxylase antizyme 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxi1+foxi2+foxi3a+foxi3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 14.5 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 2.4 | 9.5 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 2.3 | 11.5 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 1.9 | 7.5 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 1.8 | 7.1 | GO:0090244 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 1.5 | 4.6 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 1.5 | 4.4 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 1.3 | 13.5 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 1.3 | 12.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 1.2 | 3.7 | GO:0061178 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 1.1 | 6.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 1.0 | 4.8 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.9 | 8.8 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.8 | 2.4 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.8 | 19.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.8 | 10.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.7 | 9.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.7 | 18.8 | GO:0051923 | sulfation(GO:0051923) |
| 0.6 | 3.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.6 | 5.7 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.6 | 1.8 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.6 | 5.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.6 | 3.5 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.6 | 5.8 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.6 | 6.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.5 | 3.3 | GO:0043247 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.5 | 2.1 | GO:0044806 | multicellular organism aging(GO:0010259) G-quadruplex DNA unwinding(GO:0044806) |
| 0.5 | 13.2 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.5 | 1.6 | GO:0001113 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.5 | 2.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.5 | 1.5 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.5 | 11.2 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.5 | 2.8 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.5 | 6.0 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.4 | 7.2 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.4 | 5.1 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.3 | 9.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.3 | 19.7 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.3 | 2.9 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.3 | 6.9 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.3 | 40.4 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.3 | 2.5 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.3 | 1.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.3 | 3.2 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.3 | 1.8 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.3 | 3.9 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.3 | 1.8 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.3 | 1.3 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.3 | 2.8 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.2 | 4.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.2 | 7.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.2 | 14.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.2 | 4.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 4.0 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.2 | 10.8 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.2 | 3.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 2.1 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.2 | 11.9 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.2 | 5.1 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.2 | 4.7 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.2 | 2.7 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.2 | 1.1 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.2 | 5.6 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.2 | 38.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.2 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 0.8 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 6.8 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.1 | 1.5 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 | 4.5 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.1 | 2.7 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 0.7 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.1 | 0.6 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 2.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 1.4 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.1 | 20.2 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.1 | 1.9 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.1 | 0.2 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.1 | 0.3 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.1 | 2.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.3 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 1.6 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 1.6 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 4.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 6.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 3.2 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 6.9 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 1.6 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.0 | 8.7 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 4.9 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 1.4 | GO:0045197 | establishment or maintenance of apical/basal cell polarity(GO:0035088) establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) establishment or maintenance of bipolar cell polarity(GO:0061245) |
| 0.0 | 2.3 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 6.1 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 4.0 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 2.2 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 4.7 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.0 | 0.4 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.5 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.0 | 2.4 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 0.8 | GO:0006623 | protein targeting to vacuole(GO:0006623) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 | 8.0 | GO:0007409 | axonogenesis(GO:0007409) |
| 0.0 | 0.9 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 1.8 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 1.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.0 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.9 | GO:0051090 | regulation of sequence-specific DNA binding transcription factor activity(GO:0051090) |
| 0.0 | 0.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 4.9 | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter(GO:0045944) |
| 0.0 | 6.5 | GO:0007155 | cell adhesion(GO:0007155) biological adhesion(GO:0022610) |
| 0.0 | 0.5 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 1.8 | 14.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 1.5 | 11.9 | GO:0016586 | RSC complex(GO:0016586) |
| 1.3 | 8.8 | GO:0000798 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 1.2 | 9.6 | GO:0071914 | prominosome(GO:0071914) |
| 1.0 | 7.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.9 | 7.5 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.9 | 14.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.8 | 10.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.8 | 6.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.5 | 3.7 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.5 | 1.5 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.5 | 7.5 | GO:0070187 | telosome(GO:0070187) |
| 0.5 | 11.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.5 | 3.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.4 | 3.3 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.4 | 3.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.3 | 10.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 6.0 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.3 | 11.2 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.3 | 1.6 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.3 | 6.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 2.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 4.8 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.2 | 3.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 8.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 10.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 2.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 9.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 9.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 10.4 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.2 | 2.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 2.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 11.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 13.2 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.1 | 2.1 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 27.5 | GO:0005764 | lysosome(GO:0005764) |
| 0.1 | 6.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 4.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.6 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 2.9 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 3.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 7.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 4.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 5.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 10.7 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 5.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 12.8 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 3.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 59.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 12.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 4.4 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 4.3 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 3.6 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 15.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 1.6 | 9.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 1.5 | 6.0 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 1.2 | 3.6 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 1.1 | 3.2 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 1.0 | 5.1 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.9 | 7.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.9 | 2.7 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.8 | 11.2 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.7 | 14.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.6 | 13.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.6 | 17.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.6 | 1.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.6 | 4.4 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.5 | 8.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.5 | 2.7 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.4 | 3.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.4 | 10.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.4 | 3.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.4 | 1.8 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.4 | 2.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.3 | 1.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.3 | 3.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.3 | 3.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.3 | 5.6 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.3 | 1.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.3 | 4.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 3.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.3 | 9.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 3.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.3 | 6.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 3.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 6.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 7.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 4.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 18.8 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.2 | 9.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.2 | 4.8 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.2 | 2.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 10.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 1.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 0.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 3.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 8.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 6.0 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.2 | 4.0 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 15.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 11.7 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.1 | 7.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 20.2 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 2.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 1.9 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 1.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 4.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 17.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 2.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.5 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.1 | 0.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 24.0 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 56.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 2.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.9 | GO:0015370 | solute:sodium symporter activity(GO:0015370) |
| 0.0 | 1.6 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.9 | GO:0009975 | cyclase activity(GO:0009975) |
| 0.0 | 2.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.7 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 14.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.9 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 2.1 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 3.5 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 8.8 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 3.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 2.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 19.1 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.4 | 7.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.3 | 2.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 8.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 3.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 7.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.2 | 4.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 5.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.2 | 3.1 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 9.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 3.3 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 3.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 18.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.9 | 14.5 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.7 | 9.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.5 | 7.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.5 | 7.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.4 | 4.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.4 | 4.0 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.3 | 3.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.3 | 2.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 4.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 5.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.2 | 3.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 0.9 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 2.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 4.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 6.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 3.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.7 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 1.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.8 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 6.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.1 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 0.6 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 2.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.1 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |