Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
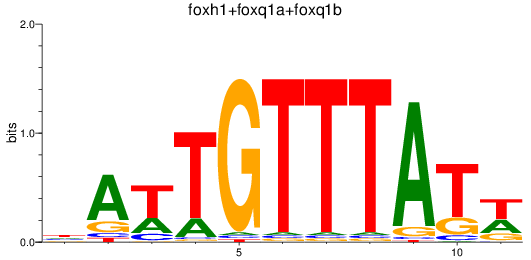
Results for foxh1+foxq1a+foxq1b
Z-value: 1.34
Transcription factors associated with foxh1+foxq1a+foxq1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxq1a
|
ENSDARG00000030896 | forkhead box Q1a |
|
foxq1b
|
ENSDARG00000055395 | forkhead box Q1b |
|
foxh1
|
ENSDARG00000055630 | forkhead box H1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxh1 | dr11_v1_chr12_-_13729263_13729263 | 0.64 | 3.4e-12 | Click! |
| foxq1b | dr11_v1_chr20_+_26683933_26683940 | 0.42 | 2.0e-05 | Click! |
| foxq1a | dr11_v1_chr2_-_722156_722170 | 0.23 | 2.8e-02 | Click! |
Activity profile of foxh1+foxq1a+foxq1b motif
Sorted Z-values of foxh1+foxq1a+foxq1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_37837245 | 41.27 |
ENSDART00000171617
|
epd
|
ependymin |
| chr17_+_15534815 | 19.29 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr4_+_8797197 | 12.37 |
ENSDART00000158671
|
sult4a1
|
sulfotransferase family 4A, member 1 |
| chr9_+_31282161 | 11.97 |
ENSDART00000010774
|
zic2a
|
zic family member 2 (odd-paired homolog, Drosophila), a |
| chr16_-_28856112 | 11.65 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr3_+_29714775 | 10.68 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr14_+_33458294 | 9.24 |
ENSDART00000075278
|
atp1b4
|
ATPase Na+/K+ transporting subunit beta 4 |
| chr6_-_9792004 | 9.17 |
ENSDART00000081129
|
cdk15
|
cyclin-dependent kinase 15 |
| chr14_-_24410673 | 9.09 |
ENSDART00000125923
|
cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr1_+_35790082 | 8.21 |
ENSDART00000085051
|
hhip
|
hedgehog interacting protein |
| chr21_+_22630627 | 7.63 |
ENSDART00000193092
|
si:dkeyp-69c1.7
|
si:dkeyp-69c1.7 |
| chr1_+_9290103 | 7.61 |
ENSDART00000055009
|
uncx4.1
|
Unc4.1 homeobox (C. elegans) |
| chr4_+_12111154 | 7.02 |
ENSDART00000036779
|
tmem178b
|
transmembrane protein 178B |
| chr21_-_32487061 | 6.90 |
ENSDART00000114359
ENSDART00000131591 ENSDART00000131477 |
si:dkeyp-72g9.4
|
si:dkeyp-72g9.4 |
| chr5_+_1278092 | 6.83 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr8_+_39634114 | 6.83 |
ENSDART00000144293
|
msi1
|
musashi RNA-binding protein 1 |
| chr7_-_26076970 | 6.69 |
ENSDART00000101120
|
zgc:92664
|
zgc:92664 |
| chr11_-_8126223 | 6.57 |
ENSDART00000091617
ENSDART00000192391 ENSDART00000101561 |
ttll7
|
tubulin tyrosine ligase-like family, member 7 |
| chr9_+_18716485 | 6.45 |
ENSDART00000135125
|
serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr1_+_24076243 | 6.42 |
ENSDART00000014608
|
mab21l2
|
mab-21-like 2 |
| chr14_+_28486213 | 6.24 |
ENSDART00000161852
|
stag2b
|
stromal antigen 2b |
| chr15_+_7992906 | 6.17 |
ENSDART00000090790
|
cadm2b
|
cell adhesion molecule 2b |
| chr2_-_55298075 | 6.15 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr20_+_33294428 | 6.15 |
ENSDART00000024104
|
mycn
|
MYCN proto-oncogene, bHLH transcription factor |
| chr16_+_31853919 | 6.14 |
ENSDART00000133886
|
atn1
|
atrophin 1 |
| chr2_-_44282796 | 5.98 |
ENSDART00000163040
ENSDART00000166923 ENSDART00000056372 ENSDART00000109251 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr22_-_29689981 | 5.91 |
ENSDART00000009223
|
pdcd4b
|
programmed cell death 4b |
| chr18_-_17001056 | 5.89 |
ENSDART00000112627
|
tph2
|
tryptophan hydroxylase 2 (tryptophan 5-monooxygenase) |
| chr15_+_23550890 | 5.89 |
ENSDART00000009796
ENSDART00000152720 |
MARK4
|
si:dkey-31m14.7 |
| chr14_-_40389699 | 5.80 |
ENSDART00000181581
ENSDART00000173398 |
pcdh19
|
protocadherin 19 |
| chr2_-_36933472 | 5.78 |
ENSDART00000170405
|
BX470229.2
|
|
| chr19_-_17526735 | 5.77 |
ENSDART00000189391
|
thrb
|
thyroid hormone receptor beta |
| chr2_+_31957554 | 5.66 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr8_-_16592491 | 5.60 |
ENSDART00000101655
|
calr
|
calreticulin |
| chr22_-_29906764 | 5.42 |
ENSDART00000019786
|
smc3
|
structural maintenance of chromosomes 3 |
| chr5_+_49744713 | 5.42 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr11_+_37612214 | 5.41 |
ENSDART00000172899
ENSDART00000077496 |
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr11_+_24046179 | 5.40 |
ENSDART00000006703
|
maf1
|
MAF1 homolog, negative regulator of RNA polymerase III |
| chr14_-_21219659 | 5.30 |
ENSDART00000089867
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr17_+_24590177 | 5.29 |
ENSDART00000092941
|
rlf
|
rearranged L-myc fusion |
| chr1_+_44711446 | 5.19 |
ENSDART00000193481
ENSDART00000003895 |
ssrp1b
|
structure specific recognition protein 1b |
| chr17_-_19022990 | 5.19 |
ENSDART00000154186
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr5_-_28767573 | 5.12 |
ENSDART00000158299
ENSDART00000043466 |
traf2a
|
Tnf receptor-associated factor 2a |
| chr7_+_28724919 | 5.12 |
ENSDART00000011324
|
ccdc102a
|
coiled-coil domain containing 102A |
| chr5_+_19712011 | 5.10 |
ENSDART00000131924
|
fam222a
|
family with sequence similarity 222, member A |
| chr3_-_49138004 | 5.08 |
ENSDART00000167173
|
gipc1
|
GIPC PDZ domain containing family, member 1 |
| chr3_+_34821327 | 5.08 |
ENSDART00000055262
|
cdk5r1a
|
cyclin-dependent kinase 5, regulatory subunit 1a (p35) |
| chr19_+_26718074 | 5.07 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr25_-_7925269 | 5.04 |
ENSDART00000014274
|
glcea
|
glucuronic acid epimerase a |
| chr17_-_42213285 | 4.99 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr16_+_25116827 | 4.88 |
ENSDART00000163244
|
si:ch211-261d7.6
|
si:ch211-261d7.6 |
| chr3_-_39696066 | 4.88 |
ENSDART00000015393
|
b9d1
|
B9 protein domain 1 |
| chr10_+_42678520 | 4.83 |
ENSDART00000182496
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr3_-_42086577 | 4.76 |
ENSDART00000083111
ENSDART00000187312 |
ttyh3a
|
tweety family member 3a |
| chr17_+_28005763 | 4.74 |
ENSDART00000155838
|
luzp1
|
leucine zipper protein 1 |
| chr1_+_53321878 | 4.71 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr2_-_44283554 | 4.60 |
ENSDART00000184684
|
mpz
|
myelin protein zero |
| chr17_-_44440832 | 4.59 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr22_+_24559947 | 4.56 |
ENSDART00000169847
|
wdr47b
|
WD repeat domain 47b |
| chr18_+_7286788 | 4.51 |
ENSDART00000022998
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr14_+_34966598 | 4.49 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr4_-_12102025 | 4.49 |
ENSDART00000048391
ENSDART00000023894 |
braf
|
B-Raf proto-oncogene, serine/threonine kinase |
| chr9_+_20554896 | 4.46 |
ENSDART00000144248
|
man1a2
|
mannosidase, alpha, class 1A, member 2 |
| chr8_-_44223473 | 4.40 |
ENSDART00000098525
|
stx2b
|
syntaxin 2b |
| chr25_+_7229046 | 4.31 |
ENSDART00000149965
ENSDART00000041820 |
lingo1a
|
leucine rich repeat and Ig domain containing 1a |
| chr1_-_53756851 | 4.27 |
ENSDART00000122445
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr2_+_24199073 | 4.22 |
ENSDART00000144110
|
map4l
|
microtubule associated protein 4 like |
| chr20_-_9963713 | 4.21 |
ENSDART00000104234
|
gjd2b
|
gap junction protein delta 2b |
| chr19_+_5143128 | 4.16 |
ENSDART00000138011
ENSDART00000108541 |
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr6_-_6423885 | 4.14 |
ENSDART00000092257
|
si:ch211-194e18.2
|
si:ch211-194e18.2 |
| chr14_-_33177935 | 4.10 |
ENSDART00000180583
ENSDART00000078856 |
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr8_-_44223899 | 4.05 |
ENSDART00000143020
|
stx2b
|
syntaxin 2b |
| chr3_-_39695856 | 4.05 |
ENSDART00000148247
|
b9d1
|
B9 protein domain 1 |
| chr13_-_27675212 | 4.03 |
ENSDART00000141035
|
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr3_+_59864872 | 4.01 |
ENSDART00000102014
|
mcrip1
|
MAPK regulated corepressor interacting protein 1 |
| chr1_+_46194333 | 3.97 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr21_+_17542473 | 3.86 |
ENSDART00000005750
ENSDART00000141326 |
stom
|
stomatin |
| chr14_-_32403554 | 3.83 |
ENSDART00000172873
ENSDART00000173408 ENSDART00000173114 ENSDART00000185594 ENSDART00000186762 ENSDART00000010982 |
fgf13a
|
fibroblast growth factor 13a |
| chr7_+_22801465 | 3.77 |
ENSDART00000052862
ENSDART00000173633 |
rbm4.1
|
RNA binding motif protein 4.1 |
| chr8_-_11324143 | 3.65 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr8_-_13210959 | 3.64 |
ENSDART00000142224
|
si:ch73-61d6.3
|
si:ch73-61d6.3 |
| chr11_+_37612672 | 3.63 |
ENSDART00000157993
|
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr2_+_8649293 | 3.47 |
ENSDART00000081442
ENSDART00000186024 |
fubp1
|
far upstream element (FUSE) binding protein 1 |
| chr12_-_34887943 | 3.42 |
ENSDART00000027379
|
bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr12_-_31422433 | 3.38 |
ENSDART00000186075
ENSDART00000153172 ENSDART00000066256 |
vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr1_+_44710955 | 3.36 |
ENSDART00000131296
ENSDART00000142187 |
ssrp1b
|
structure specific recognition protein 1b |
| chr7_+_33279108 | 3.34 |
ENSDART00000084530
|
coro2ba
|
coronin, actin binding protein, 2Ba |
| chr25_+_388258 | 3.33 |
ENSDART00000166834
|
rfx7b
|
regulatory factor X7b |
| chr23_+_6795531 | 3.30 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr3_+_22442445 | 3.29 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr24_+_39586701 | 3.28 |
ENSDART00000136447
|
si:dkey-161j23.7
|
si:dkey-161j23.7 |
| chr3_+_40289418 | 3.27 |
ENSDART00000017304
|
cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr3_+_54047342 | 3.26 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr11_-_27057572 | 3.26 |
ENSDART00000043091
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr18_+_20034023 | 3.25 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr17_-_18888959 | 3.16 |
ENSDART00000080029
|
ak7b
|
adenylate kinase 7b |
| chr18_-_34156765 | 3.14 |
ENSDART00000059400
|
c18h3orf33
|
c18h3orf33 homolog (H. sapiens) |
| chr4_+_15011341 | 3.14 |
ENSDART00000124452
|
ube2h
|
ubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast) |
| chr25_-_20024829 | 3.14 |
ENSDART00000140182
ENSDART00000174776 |
cnot4a
|
CCR4-NOT transcription complex, subunit 4a |
| chr24_+_32472155 | 3.10 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr24_+_7336807 | 3.09 |
ENSDART00000137010
|
kmt2ca
|
lysine (K)-specific methyltransferase 2Ca |
| chr4_+_75200467 | 3.01 |
ENSDART00000122593
|
CABZ01043955.1
|
|
| chr9_-_30494727 | 3.01 |
ENSDART00000148729
|
si:dkey-229b18.3
|
si:dkey-229b18.3 |
| chr7_+_30240791 | 3.00 |
ENSDART00000109243
|
sema4bb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Bb |
| chr16_-_42461263 | 2.92 |
ENSDART00000109259
|
smarcc1a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1a |
| chr17_+_25856671 | 2.92 |
ENSDART00000064817
|
wapla
|
WAPL cohesin release factor a |
| chr25_-_4717462 | 2.92 |
ENSDART00000012603
|
drd4a
|
dopamine receptor D4a |
| chr13_+_25286538 | 2.89 |
ENSDART00000180282
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr16_+_46410520 | 2.89 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr17_-_12764360 | 2.88 |
ENSDART00000003418
|
brms1la
|
breast cancer metastasis-suppressor 1-like a |
| chr6_-_19341184 | 2.88 |
ENSDART00000168236
ENSDART00000167674 |
mif4gda
|
MIF4G domain containing a |
| chr3_+_18062871 | 2.85 |
ENSDART00000112384
|
ankrd40
|
ankyrin repeat domain 40 |
| chr7_-_53117131 | 2.83 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr3_-_35554809 | 2.82 |
ENSDART00000010944
|
dctn5
|
dynactin 5 |
| chr24_+_11381400 | 2.80 |
ENSDART00000058703
|
ackr4b
|
atypical chemokine receptor 4b |
| chr10_+_20180163 | 2.76 |
ENSDART00000080016
|
ppp3ccb
|
protein phosphatase 3, catalytic subunit, gamma isozyme, b |
| chr11_+_7580079 | 2.75 |
ENSDART00000091550
ENSDART00000193223 ENSDART00000193386 |
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr6_+_33881372 | 2.74 |
ENSDART00000165710
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr7_+_29080684 | 2.73 |
ENSDART00000173709
ENSDART00000173576 |
acd
|
ACD, shelterin complex subunit and telomerase recruitment factor |
| chr24_+_26039464 | 2.72 |
ENSDART00000131017
|
tnk2a
|
tyrosine kinase, non-receptor, 2a |
| chr17_-_29902187 | 2.67 |
ENSDART00000009104
|
esrrb
|
estrogen-related receptor beta |
| chr1_+_40237276 | 2.67 |
ENSDART00000037553
|
faah2a
|
fatty acid amide hydrolase 2a |
| chr10_+_43039947 | 2.66 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr6_+_40874206 | 2.63 |
ENSDART00000004295
|
zgc:112163
|
zgc:112163 |
| chr15_+_29662401 | 2.63 |
ENSDART00000135540
|
nrip1a
|
nuclear receptor interacting protein 1a |
| chr4_-_17642168 | 2.62 |
ENSDART00000007030
|
klhl42
|
kelch-like family, member 42 |
| chr23_-_27857051 | 2.61 |
ENSDART00000043462
ENSDART00000137861 |
acvrl1
|
activin A receptor like type 1 |
| chr13_+_39182099 | 2.61 |
ENSDART00000131434
|
fam135a
|
family with sequence similarity 135, member A |
| chr16_-_21692024 | 2.61 |
ENSDART00000123597
|
si:ch211-154o6.2
|
si:ch211-154o6.2 |
| chr14_-_21218891 | 2.60 |
ENSDART00000158294
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr5_-_13766651 | 2.59 |
ENSDART00000134064
|
mxd1
|
MAX dimerization protein 1 |
| chr15_+_21712328 | 2.57 |
ENSDART00000192553
|
NKAPD1
|
zgc:162339 |
| chr1_-_45580787 | 2.57 |
ENSDART00000135089
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr8_-_7847921 | 2.49 |
ENSDART00000188094
|
zgc:113363
|
zgc:113363 |
| chr1_+_19083501 | 2.46 |
ENSDART00000002886
ENSDART00000131948 |
exosc9
|
exosome component 9 |
| chr9_+_34397516 | 2.44 |
ENSDART00000011304
ENSDART00000192973 |
med14
|
mediator complex subunit 14 |
| chr14_-_38809561 | 2.43 |
ENSDART00000159159
|
sil1
|
SIL1 nucleotide exchange factor |
| chr20_-_16156419 | 2.41 |
ENSDART00000037420
|
ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr9_+_17306162 | 2.38 |
ENSDART00000075926
|
scel
|
sciellin |
| chr3_+_34234029 | 2.35 |
ENSDART00000044859
|
znf207a
|
zinc finger protein 207, a |
| chr2_+_6126086 | 2.35 |
ENSDART00000179962
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr21_-_13225402 | 2.33 |
ENSDART00000080347
|
wdr34
|
WD repeat domain 34 |
| chr16_+_9580699 | 2.32 |
ENSDART00000165565
|
taf2
|
TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr18_+_40354998 | 2.30 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr3_-_43356082 | 2.28 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr25_+_24291156 | 2.27 |
ENSDART00000083407
|
b4galnt4a
|
beta-1,4-N-acetyl-galactosaminyl transferase 4a |
| chr8_+_4803906 | 2.26 |
ENSDART00000045533
|
tmem127
|
transmembrane protein 127 |
| chr22_+_39054012 | 2.23 |
ENSDART00000191326
|
ip6k1
|
inositol hexakisphosphate kinase 1 |
| chr12_-_28537615 | 2.22 |
ENSDART00000067762
|
MYO1D
|
si:ch211-94l19.4 |
| chr9_+_34397843 | 2.20 |
ENSDART00000146314
|
med14
|
mediator complex subunit 14 |
| chr12_+_22823219 | 2.16 |
ENSDART00000153199
ENSDART00000192571 |
afap1
|
actin filament associated protein 1 |
| chr6_+_11250316 | 2.14 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr14_-_30960470 | 2.14 |
ENSDART00000129989
|
si:ch211-191o15.6
|
si:ch211-191o15.6 |
| chr17_-_11417551 | 2.10 |
ENSDART00000128291
|
arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr2_-_6482240 | 2.07 |
ENSDART00000132623
|
rgs13
|
regulator of G protein signaling 13 |
| chr12_-_5455936 | 2.05 |
ENSDART00000109305
|
tbc1d12b
|
TBC1 domain family, member 12b |
| chr7_+_24153070 | 2.04 |
ENSDART00000076735
|
lrp10
|
low density lipoprotein receptor-related protein 10 |
| chr21_-_2707768 | 2.03 |
ENSDART00000165384
|
CABZ01101739.1
|
|
| chr12_+_18906939 | 2.03 |
ENSDART00000186074
|
josd1
|
Josephin domain containing 1 |
| chr6_+_54680730 | 1.97 |
ENSDART00000074602
|
smpd2b
|
sphingomyelin phosphodiesterase 2b, neutral membrane (neutral sphingomyelinase) |
| chr5_+_6755466 | 1.94 |
ENSDART00000163247
ENSDART00000114293 |
areg
|
amphiregulin |
| chr22_-_15437242 | 1.90 |
ENSDART00000063719
|
rpp21
|
ribonuclease P 21 subunit |
| chr10_-_20669635 | 1.89 |
ENSDART00000131361
|
kcnip3b
|
Kv channel interacting protein 3b, calsenilin |
| chr17_+_34244345 | 1.84 |
ENSDART00000006058
|
eif2s1a
|
eukaryotic translation initiation factor 2, subunit 1 alpha a |
| chr18_-_45761868 | 1.80 |
ENSDART00000025423
|
cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr23_-_41762956 | 1.79 |
ENSDART00000128302
|
stk35
|
serine/threonine kinase 35 |
| chr15_+_42285643 | 1.77 |
ENSDART00000152731
|
scaf4b
|
SR-related CTD-associated factor 4b |
| chr6_+_11250033 | 1.75 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr3_+_50172452 | 1.73 |
ENSDART00000191224
|
epn3a
|
epsin 3a |
| chr23_+_2191773 | 1.71 |
ENSDART00000190876
ENSDART00000126768 |
cc2d2a
|
coiled-coil and C2 domain containing 2A |
| chr18_+_6039141 | 1.71 |
ENSDART00000138972
|
si:ch73-386h18.1
|
si:ch73-386h18.1 |
| chr1_+_14658801 | 1.70 |
ENSDART00000192194
|
CR847936.1
|
|
| chr18_+_40355408 | 1.70 |
ENSDART00000167134
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr16_-_9675982 | 1.68 |
ENSDART00000113724
|
mal2
|
mal, T cell differentiation protein 2 (gene/pseudogene) |
| chr12_+_32729470 | 1.63 |
ENSDART00000175712
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr13_-_23956178 | 1.63 |
ENSDART00000133646
|
phactr2
|
phosphatase and actin regulator 2 |
| chr21_+_26733529 | 1.61 |
ENSDART00000168379
|
pcxa
|
pyruvate carboxylase a |
| chr11_+_24348425 | 1.59 |
ENSDART00000089747
|
nfs1
|
NFS1 cysteine desulfurase |
| chr10_-_27009413 | 1.57 |
ENSDART00000139942
ENSDART00000146983 ENSDART00000132352 |
uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr11_+_22134540 | 1.53 |
ENSDART00000190502
|
MDFI
|
si:dkey-91m3.1 |
| chr7_-_54679595 | 1.49 |
ENSDART00000165320
|
ccnd1
|
cyclin D1 |
| chr9_+_7030016 | 1.48 |
ENSDART00000148047
ENSDART00000148181 |
inpp4aa
|
inositol polyphosphate-4-phosphatase type I Aa |
| chr3_+_23488652 | 1.47 |
ENSDART00000126282
|
nr1d1
|
nuclear receptor subfamily 1, group d, member 1 |
| chr1_-_23370395 | 1.45 |
ENSDART00000143014
ENSDART00000126785 ENSDART00000159138 |
pds5a
|
PDS5 cohesin associated factor A |
| chr12_+_18906407 | 1.41 |
ENSDART00000105854
|
josd1
|
Josephin domain containing 1 |
| chr4_-_1908179 | 1.40 |
ENSDART00000139586
|
ano6
|
anoctamin 6 |
| chr13_-_31377299 | 1.39 |
ENSDART00000026692
|
ubtd1a
|
ubiquitin domain containing 1a |
| chr12_-_26609347 | 1.39 |
ENSDART00000035925
|
kctd2
|
potassium channel tetramerization domain containing 2 |
| chr18_+_13077800 | 1.37 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr14_-_15990361 | 1.33 |
ENSDART00000168075
|
trim105
|
tripartite motif containing 105 |
| chr17_+_44441042 | 1.32 |
ENSDART00000142123
|
ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr18_-_29925717 | 1.32 |
ENSDART00000099281
|
mhc2dbb
|
major histocompatibility complex class II DBB gene |
| chr7_+_24494299 | 1.30 |
ENSDART00000087568
|
nelfa
|
negative elongation factor complex member A |
| chr13_-_23956361 | 1.29 |
ENSDART00000101150
|
phactr2
|
phosphatase and actin regulator 2 |
| chr19_+_20177887 | 1.29 |
ENSDART00000008595
|
tra2a
|
transformer 2 alpha homolog |
| chr12_+_19408373 | 1.27 |
ENSDART00000114248
|
snx29
|
sorting nexin 29 |
| chr15_-_19334448 | 1.26 |
ENSDART00000062576
|
thyn1
|
thymocyte nuclear protein 1 |
| chr10_+_25355308 | 1.20 |
ENSDART00000100415
|
map3k7cl
|
map3k7 C-terminal like |
| chr15_+_32642878 | 1.18 |
ENSDART00000159932
|
trpc4b
|
transient receptor potential cation channel, subfamily C, member 4b |
| chr12_-_34716037 | 1.17 |
ENSDART00000152876
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxh1+foxq1a+foxq1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 2.0 | 12.0 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 1.5 | 6.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 1.4 | 5.8 | GO:0046552 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 1.1 | 19.2 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 1.0 | 9.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 1.0 | 5.9 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 1.0 | 2.9 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.9 | 2.7 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.8 | 2.5 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.8 | 3.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.8 | 7.9 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.7 | 3.3 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.7 | 2.6 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.6 | 6.4 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.6 | 10.7 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.6 | 5.4 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.6 | 2.9 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.6 | 1.7 | GO:0045056 | transcytosis(GO:0045056) |
| 0.5 | 3.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.5 | 9.0 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.5 | 1.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.5 | 2.8 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.5 | 1.4 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.4 | 12.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.4 | 5.0 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.4 | 4.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.4 | 3.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.4 | 2.6 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.4 | 6.9 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.4 | 3.3 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.4 | 3.3 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.4 | 6.8 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.3 | 14.6 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.3 | 41.3 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.3 | 1.5 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.3 | 8.5 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.3 | 4.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 5.1 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.3 | 12.0 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.3 | 3.8 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.3 | 5.9 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.3 | 6.9 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.3 | 1.6 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.3 | 5.1 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.2 | 5.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.2 | 7.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.2 | 6.2 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.2 | 0.7 | GO:0034035 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 1.9 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.2 | 3.2 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.2 | 1.9 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 1.6 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.2 | 0.8 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.2 | 5.1 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.2 | 4.6 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.2 | 4.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.8 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.1 | 4.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 4.8 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 5.8 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.1 | 2.0 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 7.9 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 0.5 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.1 | 0.5 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.6 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.1 | 0.6 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 2.9 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 1.3 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 4.3 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.1 | 3.6 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 2.0 | GO:0006684 | sphingomyelin metabolic process(GO:0006684) |
| 0.1 | 2.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 3.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 4.0 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 5.4 | GO:1902275 | regulation of chromatin organization(GO:1902275) |
| 0.1 | 1.6 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.4 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.1 | 1.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 11.2 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.1 | 0.4 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 2.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 11.1 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.1 | 2.9 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 2.3 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 4.9 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 3.2 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 0.8 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 0.9 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 7.6 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.0 | 1.3 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.7 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 1.0 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 3.6 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.6 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 9.4 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 3.1 | GO:0050769 | positive regulation of neurogenesis(GO:0050769) |
| 0.0 | 2.7 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.0 | 4.2 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 3.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 2.9 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 1.6 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 2.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 6.2 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.7 | GO:0045664 | regulation of neuron differentiation(GO:0045664) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.4 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 2.2 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.1 | GO:0008544 | epidermis development(GO:0008544) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.7 | GO:0016589 | NURF complex(GO:0016589) |
| 1.0 | 5.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.9 | 9.2 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.8 | 8.5 | GO:0035101 | FACT complex(GO:0035101) |
| 0.7 | 4.6 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.6 | 10.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.6 | 8.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.5 | 2.6 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.5 | 2.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.5 | 11.7 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.5 | 2.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.5 | 1.9 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.5 | 2.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.4 | 6.8 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.4 | 8.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.3 | 4.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.3 | 0.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 6.1 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.2 | 7.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 10.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 6.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 2.5 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 0.8 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.2 | 2.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 2.7 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 1.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 4.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 3.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 1.6 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 3.9 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.2 | 3.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.2 | 10.1 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 2.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 4.0 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.1 | 3.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.0 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 2.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 3.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 30.7 | GO:0005764 | lysosome(GO:0005764) |
| 0.1 | 3.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 5.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 3.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 9.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 4.1 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.1 | 3.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 4.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.7 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 11.6 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 0.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 5.6 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
| 0.0 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 3.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.7 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 2.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 2.4 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 2.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 3.8 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 2.9 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 15.7 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 1.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 6.6 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 13.0 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0043621 | protein self-association(GO:0043621) |
| 1.5 | 8.9 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 1.4 | 5.8 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 1.3 | 5.0 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 1.2 | 5.9 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 1.1 | 5.7 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.9 | 9.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.9 | 2.7 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.8 | 4.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.7 | 2.9 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.6 | 3.2 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.6 | 3.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.5 | 1.6 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.5 | 4.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.5 | 1.6 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.5 | 6.6 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.5 | 2.8 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.5 | 2.3 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.4 | 4.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.4 | 5.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.4 | 2.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.3 | 17.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.3 | 2.9 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.3 | 6.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 11.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 0.7 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 2.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 2.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 1.5 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 2.9 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.2 | 3.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 12.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 1.9 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 14.5 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.2 | 1.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 5.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 9.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 1.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 2.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 11.9 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 3.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 7.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 7.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 4.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.5 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 2.3 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 1.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 19.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 10.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 3.6 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 4.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 2.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 2.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 2.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 5.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 1.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.2 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 2.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 3.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 5.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 2.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 2.1 | GO:0019957 | chemokine binding(GO:0019956) C-C chemokine binding(GO:0019957) |
| 0.1 | 0.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.7 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 3.1 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 3.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 11.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 31.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 7.2 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 2.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 3.2 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 4.2 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 1.7 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 1.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.4 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.6 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 4.1 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 1.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 3.5 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 5.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.6 | GO:0003774 | motor activity(GO:0003774) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.3 | 5.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 7.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 1.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 5.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 2.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 5.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.2 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.9 | 2.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.6 | 5.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.5 | 4.5 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.4 | 4.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 2.9 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.3 | 4.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 2.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 9.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 4.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 2.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 4.0 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 5.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 4.6 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.1 | 1.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 4.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |