Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
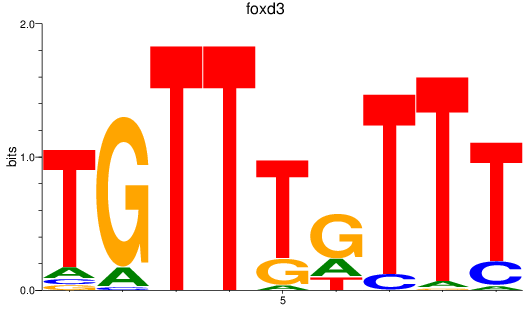
Results for foxd3
Z-value: 1.42
Transcription factors associated with foxd3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxd3
|
ENSDARG00000021032 | forkhead box D3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxd3 | dr11_v1_chr6_-_32093830_32093830 | -0.43 | 1.8e-05 | Click! |
Activity profile of foxd3 motif
Sorted Z-values of foxd3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_24559947 | 10.75 |
ENSDART00000169847
|
wdr47b
|
WD repeat domain 47b |
| chr24_+_25692802 | 10.30 |
ENSDART00000190493
|
camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr7_-_49594995 | 9.50 |
ENSDART00000174161
ENSDART00000109147 |
brsk2b
|
BR serine/threonine kinase 2b |
| chr12_+_28888975 | 9.17 |
ENSDART00000076362
|
phkg2
|
phosphorylase kinase, gamma 2 (testis) |
| chr20_-_28800999 | 8.05 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr11_+_35364445 | 7.22 |
ENSDART00000125766
|
camkvb
|
CaM kinase-like vesicle-associated b |
| chr5_+_36768674 | 7.13 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr1_+_16396870 | 7.05 |
ENSDART00000189245
ENSDART00000113266 |
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr9_+_31282161 | 6.57 |
ENSDART00000010774
|
zic2a
|
zic family member 2 (odd-paired homolog, Drosophila), a |
| chr2_+_34967022 | 6.52 |
ENSDART00000134926
|
astn1
|
astrotactin 1 |
| chr14_-_32016615 | 6.50 |
ENSDART00000105761
|
zic3
|
zic family member 3 heterotaxy 1 (odd-paired homolog, Drosophila) |
| chr18_-_23875370 | 6.37 |
ENSDART00000130163
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr6_-_30839763 | 6.18 |
ENSDART00000154228
|
sgip1a
|
SH3-domain GRB2-like (endophilin) interacting protein 1a |
| chr6_+_27146671 | 5.94 |
ENSDART00000156792
|
kif1aa
|
kinesin family member 1Aa |
| chr4_+_8797197 | 5.92 |
ENSDART00000158671
|
sult4a1
|
sulfotransferase family 4A, member 1 |
| chr1_-_50611031 | 5.85 |
ENSDART00000148285
|
ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr3_-_56924654 | 5.70 |
ENSDART00000157038
|
hid1a
|
HID1 domain containing a |
| chr4_+_11135048 | 5.66 |
ENSDART00000142389
|
cracr2ab
|
calcium release activated channel regulator 2Ab |
| chr15_+_16908085 | 5.65 |
ENSDART00000186870
|
ypel2b
|
yippee-like 2b |
| chr10_+_21789954 | 5.60 |
ENSDART00000157769
ENSDART00000171703 |
pcdh1gc5
|
protocadherin 1 gamma c 5 |
| chr4_-_13921185 | 5.59 |
ENSDART00000143202
ENSDART00000080334 |
yaf2
|
YY1 associated factor 2 |
| chr24_-_41320037 | 5.58 |
ENSDART00000129058
|
rheb
|
Ras homolog, mTORC1 binding |
| chr10_-_8032885 | 5.58 |
ENSDART00000188619
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr19_+_30633453 | 5.51 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr17_+_15433671 | 5.50 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr9_-_31278048 | 5.48 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr17_+_15433518 | 5.47 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr22_+_5574952 | 5.47 |
ENSDART00000171774
|
zgc:171566
|
zgc:171566 |
| chr10_+_17850934 | 5.46 |
ENSDART00000113666
ENSDART00000145936 |
phf24
|
PHD finger protein 24 |
| chr6_+_24817852 | 5.32 |
ENSDART00000165609
|
barhl2
|
BarH-like homeobox 2 |
| chr21_-_39081107 | 5.23 |
ENSDART00000075935
|
vtnb
|
vitronectin b |
| chr12_+_13405445 | 5.08 |
ENSDART00000089042
|
kcnh4b
|
potassium voltage-gated channel, subfamily H (eag-related), member 4b |
| chr5_-_13766651 | 4.96 |
ENSDART00000134064
|
mxd1
|
MAX dimerization protein 1 |
| chr3_+_24361096 | 4.94 |
ENSDART00000132387
|
pvalb6
|
parvalbumin 6 |
| chr11_+_24994705 | 4.86 |
ENSDART00000129211
|
zgc:92107
|
zgc:92107 |
| chr4_+_17279966 | 4.77 |
ENSDART00000067005
ENSDART00000137487 |
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr5_+_23118470 | 4.76 |
ENSDART00000149893
|
nexmifa
|
neurite extension and migration factor a |
| chr18_-_20560007 | 4.75 |
ENSDART00000141367
ENSDART00000090186 |
si:ch211-238n5.4
|
si:ch211-238n5.4 |
| chr14_+_21783229 | 4.73 |
ENSDART00000170784
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr11_+_23704410 | 4.73 |
ENSDART00000112655
|
nfasca
|
neurofascin homolog (chicken) a |
| chr8_+_21588067 | 4.72 |
ENSDART00000172190
|
ajap1
|
adherens junctions associated protein 1 |
| chr9_+_13714379 | 4.71 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr19_-_47526737 | 4.69 |
ENSDART00000186636
|
scg5
|
secretogranin V |
| chr20_-_29474859 | 4.66 |
ENSDART00000152906
ENSDART00000045249 |
scg5
|
secretogranin V |
| chr16_+_39146696 | 4.66 |
ENSDART00000121756
ENSDART00000084381 |
sybu
|
syntabulin (syntaxin-interacting) |
| chr1_+_29211540 | 4.63 |
ENSDART00000053924
|
irs2a
|
insulin receptor substrate 2a |
| chr20_-_29475172 | 4.62 |
ENSDART00000183164
|
scg5
|
secretogranin V |
| chr2_+_18988407 | 4.58 |
ENSDART00000170216
|
glula
|
glutamate-ammonia ligase (glutamine synthase) a |
| chr20_-_26042070 | 4.53 |
ENSDART00000140255
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr12_+_5081759 | 4.46 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr6_-_35446110 | 4.41 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr16_-_28856112 | 4.39 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr11_-_18323059 | 4.38 |
ENSDART00000182590
|
SFMBT1
|
Scm like with four mbt domains 1 |
| chr13_+_7292061 | 4.38 |
ENSDART00000179504
|
CABZ01072077.1
|
Danio rerio neuroblast differentiation-associated protein AHNAK-like (LOC795051), mRNA. |
| chr1_-_30039331 | 4.35 |
ENSDART00000086935
ENSDART00000143800 |
MARCH4 (1 of many)
|
zgc:153256 |
| chr18_-_23875219 | 4.30 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr16_+_47207691 | 4.28 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr23_+_19590006 | 4.28 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr13_+_38430466 | 4.26 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr8_-_34065573 | 4.24 |
ENSDART00000186946
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr16_-_24598042 | 4.23 |
ENSDART00000156407
ENSDART00000154072 |
si:dkey-56f14.4
|
si:dkey-56f14.4 |
| chr7_-_30082931 | 4.21 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr9_+_29585943 | 4.17 |
ENSDART00000185989
ENSDART00000115290 |
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr10_-_8033468 | 4.16 |
ENSDART00000140476
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr2_+_30894595 | 4.16 |
ENSDART00000132645
ENSDART00000182303 |
chmp5a
|
charged multivesicular body protein 5a |
| chr5_-_61942482 | 4.15 |
ENSDART00000097338
|
napaa
|
N-ethylmaleimide-sensitive factor attachment protein, alpha a |
| chr3_-_43356082 | 4.14 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr1_-_51606552 | 4.11 |
ENSDART00000130828
|
cnrip1a
|
cannabinoid receptor interacting protein 1a |
| chr4_-_8903240 | 4.09 |
ENSDART00000129983
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr6_+_39222598 | 4.05 |
ENSDART00000154991
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr7_-_32782430 | 4.04 |
ENSDART00000173808
|
gas2b
|
growth arrest-specific 2b |
| chr21_+_21906671 | 4.03 |
ENSDART00000016916
|
gria4b
|
glutamate receptor, ionotropic, AMPA 4b |
| chr2_-_42071558 | 4.02 |
ENSDART00000142792
|
cspp1b
|
centrosome and spindle pole associated protein 1b |
| chr23_+_17800717 | 4.01 |
ENSDART00000122654
ENSDART00000044986 |
rnd1a
|
Rho family GTPase 1a |
| chr16_+_34523515 | 4.01 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr8_+_16025554 | 4.00 |
ENSDART00000110171
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr19_-_19339285 | 4.00 |
ENSDART00000158413
ENSDART00000170479 |
cspg5b
|
chondroitin sulfate proteoglycan 5b |
| chr2_-_9059955 | 3.97 |
ENSDART00000022768
|
ak5
|
adenylate kinase 5 |
| chr22_+_12361317 | 3.95 |
ENSDART00000189963
ENSDART00000159614 |
r3hdm1
|
R3H domain containing 1 |
| chr4_+_13733838 | 3.92 |
ENSDART00000067166
ENSDART00000133157 |
cntn1b
|
contactin 1b |
| chr7_+_38770167 | 3.90 |
ENSDART00000190827
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr5_-_38384289 | 3.88 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr14_-_18672561 | 3.88 |
ENSDART00000166730
ENSDART00000006998 |
slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr8_+_41647539 | 3.87 |
ENSDART00000136492
ENSDART00000138799 ENSDART00000134404 |
si:ch211-158d24.4
|
si:ch211-158d24.4 |
| chr25_-_13789955 | 3.86 |
ENSDART00000167742
ENSDART00000165116 ENSDART00000171461 |
ckap5
|
cytoskeleton associated protein 5 |
| chr21_+_41743493 | 3.83 |
ENSDART00000192669
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr19_-_3167729 | 3.78 |
ENSDART00000110763
ENSDART00000145710 ENSDART00000074620 ENSDART00000105174 |
stm
|
starmaker |
| chr7_+_23875269 | 3.78 |
ENSDART00000101406
|
rab39bb
|
RAB39B, member RAS oncogene family b |
| chr5_-_23277939 | 3.78 |
ENSDART00000003514
|
plp1b
|
proteolipid protein 1b |
| chr7_+_60551133 | 3.77 |
ENSDART00000148038
|
lrfn4b
|
leucine rich repeat and fibronectin type III domain containing 4b |
| chr14_-_1990290 | 3.75 |
ENSDART00000183382
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr12_+_13404784 | 3.75 |
ENSDART00000167977
|
kcnh4b
|
potassium voltage-gated channel, subfamily H (eag-related), member 4b |
| chr8_+_26432677 | 3.74 |
ENSDART00000078369
ENSDART00000131925 |
zgc:136971
|
zgc:136971 |
| chr19_-_44091405 | 3.74 |
ENSDART00000132800
|
rad21b
|
RAD21 cohesin complex component b |
| chr10_+_21730585 | 3.73 |
ENSDART00000188576
|
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr23_+_19564392 | 3.73 |
ENSDART00000144746
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr2_-_12243213 | 3.72 |
ENSDART00000113081
|
gpr158b
|
G protein-coupled receptor 158b |
| chr4_-_77432218 | 3.71 |
ENSDART00000158683
|
slco1d1
|
solute carrier organic anion transporter family, member 1D1 |
| chr15_+_28202170 | 3.69 |
ENSDART00000077736
|
vtna
|
vitronectin a |
| chr8_-_16592491 | 3.65 |
ENSDART00000101655
|
calr
|
calreticulin |
| chr20_-_42203629 | 3.64 |
ENSDART00000074959
|
slc35f1
|
solute carrier family 35, member F1 |
| chr25_-_11057753 | 3.62 |
ENSDART00000186551
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr23_+_23918421 | 3.62 |
ENSDART00000046951
|
ptpn11b
|
protein tyrosine phosphatase, non-receptor type 11, b |
| chr18_-_893574 | 3.61 |
ENSDART00000150959
|
parp6a
|
poly (ADP-ribose) polymerase family, member 6a |
| chr7_+_26138240 | 3.61 |
ENSDART00000193750
ENSDART00000184942 |
nat16
|
N-acetyltransferase 16 |
| chr14_+_21783400 | 3.60 |
ENSDART00000164023
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr2_-_37043540 | 3.59 |
ENSDART00000131834
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr16_+_23811554 | 3.58 |
ENSDART00000114336
|
si:dkey-7f3.9
|
si:dkey-7f3.9 |
| chr23_+_36730713 | 3.57 |
ENSDART00000113179
|
tspan31
|
tetraspanin 31 |
| chr1_+_2190714 | 3.57 |
ENSDART00000132126
|
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr11_+_14147913 | 3.57 |
ENSDART00000022823
ENSDART00000154329 |
plppr3b
|
phospholipid phosphatase related 3b |
| chr25_+_10762016 | 3.56 |
ENSDART00000131231
|
ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr3_+_24482999 | 3.54 |
ENSDART00000059179
|
nptxra
|
neuronal pentraxin receptor a |
| chr3_-_26191960 | 3.54 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr14_-_26177156 | 3.53 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr17_-_44968177 | 3.53 |
ENSDART00000075510
|
ngb
|
neuroglobin |
| chr3_+_31925067 | 3.52 |
ENSDART00000127330
ENSDART00000055279 |
snrnp70
|
small nuclear ribonucleoprotein 70 (U1) |
| chr7_-_51546386 | 3.50 |
ENSDART00000174306
|
nhsl2
|
NHS-like 2 |
| chr9_+_21722733 | 3.48 |
ENSDART00000102021
|
sox1a
|
SRY (sex determining region Y)-box 1a |
| chr13_-_29455859 | 3.48 |
ENSDART00000077166
|
rgra
|
retinal G protein coupled receptor a |
| chr17_-_40956035 | 3.47 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr8_-_39978767 | 3.47 |
ENSDART00000083066
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr9_-_6661657 | 3.46 |
ENSDART00000133178
ENSDART00000113914 ENSDART00000061593 |
pou3f3a
|
POU class 3 homeobox 3a |
| chr2_-_37043905 | 3.44 |
ENSDART00000056514
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr17_-_44968407 | 3.43 |
ENSDART00000165850
|
ngb
|
neuroglobin |
| chr25_-_11088839 | 3.43 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr7_+_7630409 | 3.43 |
ENSDART00000172934
|
clcn3
|
chloride channel 3 |
| chr17_-_8727699 | 3.41 |
ENSDART00000049236
ENSDART00000149505 ENSDART00000148619 ENSDART00000149668 ENSDART00000148827 |
ctbp2a
|
C-terminal binding protein 2a |
| chr18_-_7137153 | 3.39 |
ENSDART00000019571
|
cd9a
|
CD9 molecule a |
| chr7_+_34487833 | 3.39 |
ENSDART00000173854
|
cln6a
|
CLN6, transmembrane ER protein a |
| chr17_-_6399920 | 3.38 |
ENSDART00000022010
|
hivep2b
|
human immunodeficiency virus type I enhancer binding protein 2b |
| chr25_+_28158352 | 3.35 |
ENSDART00000151854
|
cadps2
|
Ca++-dependent secretion activator 2 |
| chr6_-_41135215 | 3.35 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr13_-_11378355 | 3.33 |
ENSDART00000164566
|
akt3a
|
v-akt murine thymoma viral oncogene homolog 3a |
| chr20_-_9462433 | 3.32 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr17_-_16133249 | 3.32 |
ENSDART00000030919
|
pnoca
|
prepronociceptin a |
| chr3_-_22191132 | 3.30 |
ENSDART00000154226
ENSDART00000155528 ENSDART00000155190 |
maptb
|
microtubule-associated protein tau b |
| chr21_+_11415224 | 3.30 |
ENSDART00000049036
|
zgc:92275
|
zgc:92275 |
| chr21_+_25181003 | 3.30 |
ENSDART00000169700
|
si:dkey-183i3.9
|
si:dkey-183i3.9 |
| chr15_-_21877726 | 3.27 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr13_+_1100197 | 3.25 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr2_+_24177006 | 3.24 |
ENSDART00000132582
|
map4l
|
microtubule associated protein 4 like |
| chr10_+_26515946 | 3.24 |
ENSDART00000134276
|
synj1
|
synaptojanin 1 |
| chr5_+_19314574 | 3.24 |
ENSDART00000133247
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr8_-_25566347 | 3.23 |
ENSDART00000138289
ENSDART00000078022 |
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr19_-_47527093 | 3.22 |
ENSDART00000171379
ENSDART00000171140 ENSDART00000114886 |
scg5
|
secretogranin V |
| chr2_+_5563077 | 3.22 |
ENSDART00000111220
|
mb21d2
|
Mab-21 domain containing 2 |
| chr3_-_27880229 | 3.21 |
ENSDART00000151404
|
abat
|
4-aminobutyrate aminotransferase |
| chr3_+_54012708 | 3.21 |
ENSDART00000154542
|
olfm2a
|
olfactomedin 2a |
| chr2_+_21128391 | 3.19 |
ENSDART00000136814
|
pip4k2ab
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha b |
| chr19_-_9503473 | 3.18 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr14_-_32403554 | 3.17 |
ENSDART00000172873
ENSDART00000173408 ENSDART00000173114 ENSDART00000185594 ENSDART00000186762 ENSDART00000010982 |
fgf13a
|
fibroblast growth factor 13a |
| chr1_-_22834824 | 3.14 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr9_-_33785093 | 3.12 |
ENSDART00000140779
ENSDART00000059837 |
fundc1
|
FUN14 domain containing 1 |
| chr7_-_26087807 | 3.10 |
ENSDART00000052989
|
ache
|
acetylcholinesterase |
| chr3_-_37758487 | 3.09 |
ENSDART00000150938
|
si:dkey-260c8.6
|
si:dkey-260c8.6 |
| chr1_+_35790082 | 3.08 |
ENSDART00000085051
|
hhip
|
hedgehog interacting protein |
| chr11_-_29082429 | 3.08 |
ENSDART00000041443
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr22_+_20208185 | 3.08 |
ENSDART00000142748
|
si:dkey-110c1.7
|
si:dkey-110c1.7 |
| chr3_-_29910547 | 3.07 |
ENSDART00000151501
|
RUNDC1
|
si:dkey-151m15.5 |
| chr6_-_9792004 | 3.05 |
ENSDART00000081129
|
cdk15
|
cyclin-dependent kinase 15 |
| chr14_-_18671334 | 3.04 |
ENSDART00000182381
|
slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr8_-_52413032 | 3.04 |
ENSDART00000183039
|
CABZ01070469.1
|
|
| chr7_+_61050329 | 3.04 |
ENSDART00000115355
|
nwd2
|
NACHT and WD repeat domain containing 2 |
| chr5_-_29122615 | 3.03 |
ENSDART00000144802
|
whrnb
|
whirlin b |
| chr21_-_31143903 | 3.02 |
ENSDART00000111571
|
rap1gap2b
|
RAP1 GTPase activating protein 2b |
| chr4_-_17629444 | 3.02 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr14_-_16082806 | 3.00 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr8_+_46418996 | 3.00 |
ENSDART00000144285
|
si:ch211-196g2.4
|
si:ch211-196g2.4 |
| chr9_+_29589790 | 2.99 |
ENSDART00000140388
|
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr7_+_52712807 | 2.98 |
ENSDART00000174095
ENSDART00000174377 ENSDART00000174061 ENSDART00000174094 ENSDART00000110906 ENSDART00000174071 ENSDART00000174238 |
znf280d
|
zinc finger protein 280D |
| chr7_+_33314925 | 2.98 |
ENSDART00000148590
|
coro2ba
|
coronin, actin binding protein, 2Ba |
| chr17_-_7818944 | 2.97 |
ENSDART00000135538
ENSDART00000037541 |
rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr9_-_33477588 | 2.96 |
ENSDART00000144150
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr14_+_31618982 | 2.96 |
ENSDART00000026195
|
slc9a6a
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6a |
| chr9_+_44430974 | 2.96 |
ENSDART00000056846
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr10_-_34772211 | 2.95 |
ENSDART00000145450
ENSDART00000134307 |
dclk1a
|
doublecortin-like kinase 1a |
| chr14_-_33454595 | 2.95 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr14_+_34966598 | 2.94 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr6_-_53426773 | 2.89 |
ENSDART00000162791
|
mst1
|
macrophage stimulating 1 |
| chr3_-_30123113 | 2.88 |
ENSDART00000153562
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr7_-_35708450 | 2.87 |
ENSDART00000193886
|
irx5a
|
iroquois homeobox 5a |
| chr14_-_2348917 | 2.86 |
ENSDART00000159004
|
si:ch73-233f7.8
|
si:ch73-233f7.8 |
| chr2_-_36933472 | 2.84 |
ENSDART00000170405
|
BX470229.2
|
|
| chr5_-_38384755 | 2.84 |
ENSDART00000188573
ENSDART00000051233 |
mink1
|
misshapen-like kinase 1 |
| chr11_-_17713987 | 2.84 |
ENSDART00000090401
|
fam19a4b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4b |
| chr1_-_22757145 | 2.84 |
ENSDART00000134719
|
prom1b
|
prominin 1 b |
| chr25_-_31763897 | 2.83 |
ENSDART00000041740
|
ubl7a
|
ubiquitin-like 7a (bone marrow stromal cell-derived) |
| chr13_+_24022963 | 2.82 |
ENSDART00000028285
|
pgbd5
|
piggyBac transposable element derived 5 |
| chr11_+_30817943 | 2.80 |
ENSDART00000150130
ENSDART00000159997 |
cacna1ab
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, b |
| chr4_+_10365857 | 2.79 |
ENSDART00000138890
|
kcnd2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr23_-_14403939 | 2.78 |
ENSDART00000090930
|
nkain4
|
sodium/potassium transporting ATPase interacting 4 |
| chr15_+_40079468 | 2.78 |
ENSDART00000154947
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr9_+_2574122 | 2.78 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr12_-_19346678 | 2.76 |
ENSDART00000044860
|
maff
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr6_-_40922971 | 2.76 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr20_-_39103119 | 2.75 |
ENSDART00000143379
|
rcan2
|
regulator of calcineurin 2 |
| chr10_+_17936441 | 2.74 |
ENSDART00000146489
ENSDART00000064866 |
prkab1a
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit, a |
| chr23_-_36670369 | 2.73 |
ENSDART00000006881
|
zbtb39
|
zinc finger and BTB domain containing 39 |
| chr5_+_61361815 | 2.73 |
ENSDART00000009507
|
gatsl2
|
GATS protein-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxd3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.4 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 1.7 | 12.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 1.5 | 4.5 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 1.4 | 6.8 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 1.4 | 5.5 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 1.2 | 4.8 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 1.2 | 4.8 | GO:2000048 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 1.2 | 4.7 | GO:0060074 | synapse maturation(GO:0060074) |
| 1.1 | 4.4 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 1.0 | 4.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 1.0 | 5.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 1.0 | 3.9 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.9 | 2.8 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.9 | 2.7 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.9 | 2.7 | GO:1903792 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.8 | 5.8 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.8 | 3.1 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.7 | 3.0 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.7 | 2.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.7 | 2.0 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.7 | 2.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.7 | 2.0 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.7 | 5.9 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.7 | 3.3 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.6 | 3.9 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.6 | 3.2 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.6 | 1.9 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.6 | 2.5 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.6 | 1.9 | GO:0019852 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.6 | 1.9 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.6 | 2.4 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.6 | 1.8 | GO:0044060 | regulation of endocrine process(GO:0044060) endocrine hormone secretion(GO:0060986) |
| 0.6 | 8.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.6 | 24.3 | GO:0046883 | regulation of hormone secretion(GO:0046883) |
| 0.6 | 1.7 | GO:0045830 | regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.6 | 5.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.6 | 1.7 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.5 | 3.8 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.5 | 1.6 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.5 | 2.1 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.5 | 2.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.5 | 2.6 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.5 | 1.5 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.5 | 2.5 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.5 | 2.0 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.5 | 2.0 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.5 | 3.4 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.5 | 3.8 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.5 | 2.4 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.5 | 6.7 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.5 | 1.9 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.5 | 3.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.5 | 1.4 | GO:1903646 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.5 | 1.4 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.5 | 0.5 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.4 | 2.7 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.4 | 2.2 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.4 | 2.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.4 | 2.6 | GO:0050951 | detection of temperature stimulus(GO:0016048) sensory perception of temperature stimulus(GO:0050951) |
| 0.4 | 5.6 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.4 | 1.3 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.4 | 3.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.4 | 7.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.4 | 4.0 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.4 | 1.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.4 | 2.8 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.4 | 2.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.4 | 3.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.4 | 3.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.4 | 1.9 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.4 | 1.9 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.4 | 1.1 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.4 | 1.1 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.4 | 4.9 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.4 | 1.1 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.4 | 2.2 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.4 | 3.7 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.4 | 11.8 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.4 | 6.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.4 | 2.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.4 | 1.8 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.4 | 1.8 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.3 | 1.4 | GO:0035522 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.3 | 1.7 | GO:1901906 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.3 | 2.1 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.3 | 1.4 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.3 | 1.0 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.3 | 2.0 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.3 | 4.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.3 | 0.9 | GO:0021698 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.3 | 1.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 5.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.3 | 1.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.3 | 2.0 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.3 | 5.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.3 | 1.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.3 | 1.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.3 | 4.8 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.3 | 2.2 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.3 | 0.8 | GO:0039529 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.3 | 5.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.3 | 2.5 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.3 | 1.4 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.3 | 3.0 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 1.3 | GO:0043584 | nose development(GO:0043584) |
| 0.3 | 2.4 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.3 | 1.6 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.3 | 1.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.3 | 1.5 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.3 | 1.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.3 | 0.8 | GO:0001120 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.2 | 1.7 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.2 | 2.5 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.2 | 2.2 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.2 | 0.7 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.2 | 2.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 2.8 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.2 | 0.7 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.2 | 2.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 6.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 6.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.2 | 1.1 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.2 | 4.1 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.2 | 2.2 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.2 | 2.0 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.2 | 1.6 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 0.7 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.2 | 0.7 | GO:0031645 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.2 | 0.9 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.2 | 4.0 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 2.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 1.3 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.2 | 4.9 | GO:0042670 | retinal cone cell differentiation(GO:0042670) |
| 0.2 | 5.1 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.2 | 3.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.2 | 1.7 | GO:0000423 | macromitophagy(GO:0000423) |
| 0.2 | 1.3 | GO:0061316 | canonical Wnt signaling pathway involved in heart development(GO:0061316) |
| 0.2 | 1.0 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.2 | 1.0 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.2 | 2.0 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.2 | 3.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 0.4 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.2 | 1.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.2 | 1.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.2 | 6.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 1.9 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.2 | 0.8 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.2 | 1.3 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 0.8 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 | 2.5 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.2 | 4.7 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.2 | 1.3 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 | 3.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 0.6 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.2 | 2.6 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.2 | 3.1 | GO:0050685 | positive regulation of mRNA processing(GO:0050685) |
| 0.2 | 2.0 | GO:0043536 | positive regulation of blood vessel endothelial cell migration(GO:0043536) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 0.7 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 0.5 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.2 | 1.6 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.2 | 1.6 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.2 | 0.9 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.2 | 1.6 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 | 2.4 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.2 | 4.6 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.2 | 1.2 | GO:0051382 | kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.2 | 2.6 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.2 | 7.0 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.2 | 1.6 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.2 | 4.4 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.2 | 0.8 | GO:0003311 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.2 | 0.6 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.2 | 0.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 0.8 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 0.9 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) quinolinate metabolic process(GO:0046874) |
| 0.2 | 1.5 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.2 | 1.4 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 2.6 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 0.7 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.6 | GO:0042308 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.1 | 4.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 9.3 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.1 | 1.1 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.1 | 6.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 30.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.3 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.6 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.9 | GO:0042311 | vasodilation(GO:0042311) |
| 0.1 | 0.5 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.1 | 7.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 3.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 3.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 2.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 4.9 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.8 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.1 | 1.3 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 2.2 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 | 4.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 1.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.9 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 5.7 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.1 | 0.7 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 1.6 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 10.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.2 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.6 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.6 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.7 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 3.7 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 4.1 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.1 | 0.3 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.1 | 4.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 1.4 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 1.0 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 2.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 1.7 | GO:1903288 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 0.8 | GO:0070206 | protein trimerization(GO:0070206) protein homotrimerization(GO:0070207) |
| 0.1 | 0.8 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.6 | GO:0072425 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.1 | 2.6 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 1.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 2.2 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.6 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 6.8 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.1 | 1.2 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.1 | 0.9 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 3.3 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.1 | 1.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.4 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 2.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 1.9 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 2.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.7 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.5 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.1 | 1.3 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 | 0.6 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 2.0 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.1 | 5.0 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 0.8 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.5 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 11.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 2.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.4 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 1.4 | GO:0009411 | response to UV(GO:0009411) |
| 0.1 | 3.8 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 0.3 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.1 | 0.3 | GO:0009258 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 1.0 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 6.8 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 1.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.8 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 8.8 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.1 | 0.7 | GO:0030656 | retinoic acid biosynthetic process(GO:0002138) regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 0.6 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 1.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 3.6 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.1 | 1.9 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.1 | 0.6 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 1.6 | GO:0043153 | entrainment of circadian clock(GO:0009649) entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 1.9 | GO:0003401 | axis elongation(GO:0003401) |
| 0.1 | 1.4 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.9 | GO:0033505 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.1 | 0.5 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 1.0 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 1.2 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.1 | 0.2 | GO:0036315 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.6 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 1.6 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 1.0 | GO:0043506 | activation of JUN kinase activity(GO:0007257) regulation of JUN kinase activity(GO:0043506) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 1.4 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.1 | 3.4 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 1.1 | GO:0001736 | establishment of planar polarity(GO:0001736) establishment of tissue polarity(GO:0007164) |
| 0.1 | 1.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 1.0 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 1.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.8 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 0.2 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.1 | 0.5 | GO:0031529 | ruffle organization(GO:0031529) ruffle assembly(GO:0097178) |
| 0.1 | 0.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 2.2 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.3 | GO:1903826 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.8 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 0.8 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 0.5 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 0.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 9.6 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 1.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.7 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.1 | 2.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 1.1 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.1 | 1.3 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.9 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 6.8 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 0.7 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 1.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.8 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.0 | 0.7 | GO:0015833 | peptide transport(GO:0015833) |
| 0.0 | 0.3 | GO:0031057 | negative regulation of histone modification(GO:0031057) regulation of histone acetylation(GO:0035065) |
| 0.0 | 2.9 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 0.6 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 1.6 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
| 0.0 | 0.4 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.6 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.8 | GO:0055069 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 0.7 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 2.6 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 0.8 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.6 | GO:0050906 | detection of stimulus involved in sensory perception(GO:0050906) |
| 0.0 | 5.6 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.1 | GO:0032239 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 2.1 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.8 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.7 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 1.3 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 7.1 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.6 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 2.9 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 3.9 | GO:0030900 | forebrain development(GO:0030900) |
| 0.0 | 4.3 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.0 | 0.6 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 1.9 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.1 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.7 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.3 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 1.6 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.5 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.6 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 0.4 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.8 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.5 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.0 | 1.7 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 4.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.5 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 1.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.5 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 2.5 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.1 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 8.3 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 1.4 | GO:0048565 | digestive tract development(GO:0048565) |
| 0.0 | 1.3 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 3.8 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.5 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.4 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.5 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.3 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.0 | 1.4 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.6 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 0.2 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.5 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.4 | GO:0048753 | pigment granule organization(GO:0048753) |
| 0.0 | 0.9 | GO:0031929 | TOR signaling(GO:0031929) |
| 0.0 | 0.3 | GO:0072665 | protein localization to vacuole(GO:0072665) |
| 0.0 | 1.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.4 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.7 | GO:0071219 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 1.4 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 1.2 | GO:0006402 | mRNA catabolic process(GO:0006402) |
| 0.0 | 1.0 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.3 | GO:0035924 | cellular response to vascular endothelial growth factor stimulus(GO:0035924) |
| 0.0 | 0.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.7 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.7 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.5 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.7 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 1.1 | 5.7 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 1.1 | 4.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 1.0 | 4.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.8 | 8.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.8 | 9.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.7 | 4.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.7 | 3.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.7 | 6.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.6 | 1.9 | GO:0031213 | RSF complex(GO:0031213) |
| 0.6 | 2.5 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.6 | 2.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.6 | 3.7 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.5 | 4.4 | GO:0071914 | prominosome(GO:0071914) |
| 0.5 | 3.7 | GO:0030892 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.5 | 1.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.5 | 2.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.5 | 2.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.5 | 8.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.5 | 1.4 | GO:0072380 | TRC complex(GO:0072380) |
| 0.4 | 1.7 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.4 | 5.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.4 | 2.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.4 | 1.8 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.4 | 15.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.4 | 2.9 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.4 | 3.2 | GO:0032426 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.3 | 2.8 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.3 | 2.7 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.3 | 1.6 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.3 | 1.6 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.3 | 2.5 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.3 | 1.2 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.3 | 2.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.3 | 0.9 | GO:1990745 | EARP complex(GO:1990745) |
| 0.3 | 1.5 | GO:0000938 | GARP complex(GO:0000938) |
| 0.3 | 6.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.3 | 4.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.3 | 2.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.3 | 21.9 | GO:0030141 | secretory granule(GO:0030141) |
| 0.3 | 4.0 | GO:0070187 | telosome(GO:0070187) |
| 0.3 | 1.9 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.3 | 1.0 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.3 | 2.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.3 | 1.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.3 | 2.0 | GO:0016586 | RSC complex(GO:0016586) |
| 0.3 | 3.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 1.5 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.2 | 3.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 0.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 5.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.2 | 2.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 1.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 3.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 3.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 8.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 1.4 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.2 | 1.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 4.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.2 | 1.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.2 | 11.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 2.0 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.2 | 1.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 0.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 1.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 1.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 0.6 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.2 | 3.7 | GO:0071010 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.2 | 6.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 6.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 5.3 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.2 | 2.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 0.8 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.6 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 1.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.7 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 2.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.0 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 37.8 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 3.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.1 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.1 | 0.9 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 3.9 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 4.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 6.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.3 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 1.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 2.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 1.0 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 0.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 8.6 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 11.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 1.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.9 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.7 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 3.0 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 5.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.4 | GO:0043679 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.1 | 0.6 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 1.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 2.6 | GO:0032590 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.1 | 2.4 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.8 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.5 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 2.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 3.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.4 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 15.9 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 1.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 1.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 3.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 2.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 1.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 4.9 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 1.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.5 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 3.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 6.1 | GO:0070382 | exocytic vesicle(GO:0070382) secretory vesicle(GO:0099503) |
| 0.1 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.0 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 0.5 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 2.8 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.1 | 0.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 3.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 3.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 2.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.0 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 6.3 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 6.8 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 2.6 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 17.1 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 2.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.4 | GO:0005773 | vacuole(GO:0005773) |
| 0.0 | 5.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.7 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 3.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 9.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 1.7 | 10.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 1.4 | 11.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 1.2 | 4.8 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 1.0 | 6.7 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.9 | 3.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.9 | 2.6 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.8 | 5.8 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.8 | 2.5 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.8 | 16.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.7 | 2.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.7 | 4.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.7 | 2.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.7 | 10.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.7 | 3.3 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.6 | 1.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.5 | 2.1 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.5 | 1.6 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.5 | 3.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.5 | 2.5 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.5 | 2.0 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.5 | 2.5 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.5 | 2.0 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.5 | 1.4 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.5 | 2.8 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.5 | 2.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.4 | 3.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 6.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.4 | 3.5 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.4 | 1.3 | GO:0052855 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.4 | 2.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.4 | 1.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.4 | 1.3 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.4 | 1.2 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.4 | 9.3 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.4 | 8.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.4 | 5.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.4 | 4.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.4 | 1.9 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.4 | 5.3 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.4 | 1.5 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.4 | 4.8 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.4 | 1.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.4 | 1.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.4 | 2.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.4 | 1.8 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.3 | 1.7 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.3 | 1.4 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.3 | 8.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 1.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.3 | 1.9 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.3 | 0.9 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.3 | 2.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.3 | 4.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.3 | 3.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.3 | 1.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.3 | 4.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.3 | 1.2 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.3 | 5.5 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.3 | 2.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 1.4 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.3 | 3.8 | GO:0034596 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) |
| 0.3 | 1.0 | GO:0032028 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.3 | 1.3 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.3 | 1.0 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.3 | 3.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.3 | 0.8 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.3 | 1.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.3 | 1.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.3 | 1.3 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.3 | 3.3 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.3 | 3.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 2.0 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.2 | 1.0 | GO:0030626 | U12 snRNA binding(GO:0030626) pre-mRNA intronic binding(GO:0097157) |
| 0.2 | 2.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.2 | 2.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 0.7 | GO:0042166 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 0.2 | 6.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 8.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 1.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 7.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 0.8 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 3.4 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.2 | 1.9 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.2 | 3.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.0 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 1.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 1.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 1.3 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.2 | 2.6 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 1.3 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.2 | 7.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 0.5 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 9.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.2 | 0.5 | GO:0009384 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.2 | 4.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 1.7 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.2 | 0.6 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 3.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.2 | 4.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 16.9 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.2 | 1.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.2 | 0.8 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.2 | 5.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 2.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 0.9 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.4 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.1 | 1.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 6.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 3.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 2.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 1.2 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.4 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.1 | 0.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.5 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 2.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 1.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 3.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 1.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 3.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 2.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 1.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.5 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 3.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 2.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 4.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 15.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 3.6 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 1.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.6 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 3.7 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 5.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 4.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 36.6 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 1.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 1.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 2.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 4.1 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 0.7 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 1.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.3 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 2.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 6.8 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.0 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 1.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.8 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 1.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 3.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 2.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 2.6 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 1.4 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 2.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.3 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.1 | 0.8 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 1.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 3.1 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 0.6 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 1.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 1.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.7 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.4 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 3.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 0.5 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.5 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.7 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 1.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 6.5 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.0 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 1.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 1.0 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 3.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.0 | 0.0 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.0 | 15.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.4 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 1.8 | GO:0015216 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 2.2 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 2.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.0 | 1.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.0 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 5.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 1.0 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 11.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.6 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 1.2 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 2.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.7 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.7 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.8 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 1.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 6.8 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 2.4 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.3 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 1.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 2.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 9.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 0.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.5 | 1.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 22.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.3 | 11.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 2.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 1.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 2.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 3.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.2 | 1.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 1.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 0.5 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.2 | 8.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.2 | 1.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 2.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 1.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 2.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 3.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 0.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 2.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 2.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 3.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 2.9 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.0 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 3.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 2.5 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 3.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 1.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 0.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 3.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 0.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.8 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 1.2 | 7.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.7 | 9.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.5 | 8.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.4 | 3.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.4 | 3.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 1.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.3 | 1.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.3 | 3.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.3 | 4.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 2.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.2 | 4.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 3.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 1.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.2 | 2.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 6.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 0.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 2.0 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 1.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 2.0 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 8.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 0.7 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.2 | 12.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 3.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 2.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 1.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 3.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 3.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 3.7 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 0.6 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 1.9 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 2.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 1.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 4.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.8 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 2.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 0.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 0.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 0.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 0.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.5 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.3 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.8 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |
| 0.0 | 0.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |