Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
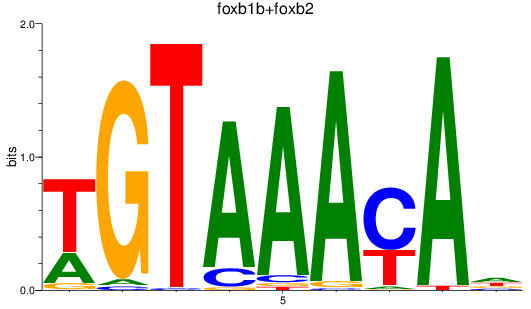
Results for foxb1b+foxb2
Z-value: 1.29
Transcription factors associated with foxb1b+foxb2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxb2
|
ENSDARG00000037475 | forkhead box B2 |
|
foxb1b
|
ENSDARG00000053650 | forkhead box B1b |
|
foxb1b
|
ENSDARG00000110408 | forkhead box B1b |
|
foxb1b
|
ENSDARG00000113373 | forkhead box B1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxb1b | dr11_v1_chr7_+_29461060_29461060 | 0.68 | 3.5e-14 | Click! |
| foxb2 | dr11_v1_chr8_-_38506339_38506339 | 0.61 | 8.4e-11 | Click! |
Activity profile of foxb1b+foxb2 motif
Sorted Z-values of foxb1b+foxb2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_19953089 | 18.22 |
ENSDART00000153828
|
atp2b3b
|
ATPase plasma membrane Ca2+ transporting 3b |
| chr23_-_27633730 | 10.51 |
ENSDART00000103639
|
arf3a
|
ADP-ribosylation factor 3a |
| chr6_+_36942966 | 10.24 |
ENSDART00000028895
|
negr1
|
neuronal growth regulator 1 |
| chr20_-_34801181 | 9.87 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr4_+_8797197 | 9.80 |
ENSDART00000158671
|
sult4a1
|
sulfotransferase family 4A, member 1 |
| chr12_+_28367557 | 9.75 |
ENSDART00000066294
|
cdk5r1b
|
cyclin-dependent kinase 5, regulatory subunit 1b (p35) |
| chr14_-_7885707 | 9.34 |
ENSDART00000029981
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr14_-_26177156 | 9.32 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr19_+_10396042 | 9.25 |
ENSDART00000028048
ENSDART00000151735 |
necap1
|
NECAP endocytosis associated 1 |
| chr23_-_29502287 | 8.89 |
ENSDART00000141075
ENSDART00000053807 |
kif1b
|
kinesin family member 1B |
| chr6_-_9792004 | 8.88 |
ENSDART00000081129
|
cdk15
|
cyclin-dependent kinase 15 |
| chr16_-_43025885 | 8.77 |
ENSDART00000193146
ENSDART00000157302 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr16_-_12173554 | 8.73 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr21_+_11684830 | 8.28 |
ENSDART00000147473
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr15_+_37105986 | 8.20 |
ENSDART00000157762
|
aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr2_-_8017579 | 8.13 |
ENSDART00000040209
|
ephb3a
|
eph receptor B3a |
| chr1_+_16127825 | 7.99 |
ENSDART00000122503
|
tusc3
|
tumor suppressor candidate 3 |
| chr2_-_24462277 | 7.85 |
ENSDART00000033922
|
kcnn1a
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1a |
| chr2_+_24177006 | 7.53 |
ENSDART00000132582
|
map4l
|
microtubule associated protein 4 like |
| chr14_-_49063157 | 7.11 |
ENSDART00000021260
|
sept8b
|
septin 8b |
| chr16_+_46000956 | 6.97 |
ENSDART00000101753
ENSDART00000162393 |
mtmr11
|
myotubularin related protein 11 |
| chr21_+_11685009 | 6.96 |
ENSDART00000014668
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr24_-_4973765 | 6.95 |
ENSDART00000127597
|
zic1
|
zic family member 1 (odd-paired homolog, Drosophila) |
| chr9_-_44295071 | 6.90 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr3_+_19207176 | 6.86 |
ENSDART00000087803
|
rln3a
|
relaxin 3a |
| chr2_+_29976419 | 6.85 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr1_-_28749604 | 6.85 |
ENSDART00000148522
|
GK3P
|
zgc:172295 |
| chr8_-_25120231 | 6.84 |
ENSDART00000147308
|
amigo1
|
adhesion molecule with Ig-like domain 1 |
| chr14_-_17599452 | 6.83 |
ENSDART00000080042
|
rab33a
|
RAB33A, member RAS oncogene family |
| chr21_+_30563115 | 6.67 |
ENSDART00000028566
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr4_+_17280868 | 6.58 |
ENSDART00000145349
|
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr13_+_1100197 | 6.51 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr1_+_53954230 | 6.50 |
ENSDART00000037729
ENSDART00000159900 |
ccsapa
|
centriole, cilia and spindle-associated protein a |
| chr18_-_14734678 | 6.40 |
ENSDART00000142462
|
tshz3a
|
teashirt zinc finger homeobox 3a |
| chr10_-_15405564 | 6.38 |
ENSDART00000020665
|
sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr1_+_25801648 | 6.35 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr6_-_40744720 | 6.33 |
ENSDART00000154916
ENSDART00000186922 |
p4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr17_-_20287530 | 6.31 |
ENSDART00000078703
ENSDART00000191289 |
add3b
|
adducin 3 (gamma) b |
| chr14_-_34044369 | 6.25 |
ENSDART00000149396
ENSDART00000123607 ENSDART00000190746 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr20_-_19590378 | 6.22 |
ENSDART00000152588
|
baalcb
|
brain and acute leukemia, cytoplasmic b |
| chr19_+_6938289 | 6.14 |
ENSDART00000139122
ENSDART00000178832 |
flot1b
|
flotillin 1b |
| chr14_-_25599002 | 6.11 |
ENSDART00000040955
|
slc25a48
|
solute carrier family 25, member 48 |
| chr7_+_7019911 | 6.11 |
ENSDART00000172421
|
rbm14b
|
RNA binding motif protein 14b |
| chr25_-_15049694 | 6.10 |
ENSDART00000162485
ENSDART00000164384 ENSDART00000165632 ENSDART00000159490 |
pax6a
|
paired box 6a |
| chr6_-_32999646 | 6.06 |
ENSDART00000159510
|
adcyap1r1b
|
adenylate cyclase activating polypeptide 1b (pituitary) receptor type I |
| chr6_+_23752593 | 6.06 |
ENSDART00000164366
|
zgc:158654
|
zgc:158654 |
| chr11_-_29833698 | 6.03 |
ENSDART00000079149
|
xk
|
X-linked Kx blood group (McLeod syndrome) |
| chr11_-_4235811 | 5.96 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr18_+_24922125 | 5.91 |
ENSDART00000180385
|
rgma
|
repulsive guidance molecule family member a |
| chr12_+_31729075 | 5.89 |
ENSDART00000152973
|
RNF157
|
si:dkey-49c17.3 |
| chr20_+_25486206 | 5.89 |
ENSDART00000172076
|
hook1
|
hook microtubule-tethering protein 1 |
| chr10_+_25219728 | 5.82 |
ENSDART00000193829
|
grm5a
|
glutamate receptor, metabotropic 5a |
| chr12_-_19346678 | 5.78 |
ENSDART00000044860
|
maff
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr3_+_34821327 | 5.72 |
ENSDART00000055262
|
cdk5r1a
|
cyclin-dependent kinase 5, regulatory subunit 1a (p35) |
| chr17_-_40397752 | 5.69 |
ENSDART00000178483
|
BX548062.1
|
|
| chr22_+_27090136 | 5.64 |
ENSDART00000136770
|
si:dkey-246e1.3
|
si:dkey-246e1.3 |
| chr3_-_26341959 | 5.63 |
ENSDART00000169344
ENSDART00000142878 ENSDART00000087196 |
zgc:153240
|
zgc:153240 |
| chr6_+_40671336 | 5.61 |
ENSDART00000111639
ENSDART00000186617 |
rereb
|
arginine-glutamic acid dipeptide (RE) repeats b |
| chr7_-_32782430 | 5.57 |
ENSDART00000173808
|
gas2b
|
growth arrest-specific 2b |
| chr6_-_13187168 | 5.54 |
ENSDART00000193286
ENSDART00000188350 ENSDART00000150036 ENSDART00000149940 |
adam23a
|
ADAM metallopeptidase domain 23a |
| chr7_+_48288762 | 5.52 |
ENSDART00000083569
|
oaz2b
|
ornithine decarboxylase antizyme 2b |
| chr7_-_31941670 | 5.42 |
ENSDART00000180929
ENSDART00000075389 |
bdnf
|
brain-derived neurotrophic factor |
| chr6_-_30658755 | 5.41 |
ENSDART00000065215
ENSDART00000181302 |
lurap1
|
leucine rich adaptor protein 1 |
| chr21_+_20901505 | 5.40 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr23_-_3703569 | 5.31 |
ENSDART00000143731
|
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr7_+_48460239 | 5.26 |
ENSDART00000052113
|
lingo1b
|
leucine rich repeat and Ig domain containing 1b |
| chr2_+_24177190 | 5.25 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr21_-_29100110 | 5.19 |
ENSDART00000142598
|
timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr21_-_30284404 | 5.18 |
ENSDART00000066363
|
zgc:175066
|
zgc:175066 |
| chr19_-_28367413 | 5.14 |
ENSDART00000079092
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr2_+_26240631 | 5.11 |
ENSDART00000129895
|
palm1b
|
paralemmin 1b |
| chr13_+_32446169 | 5.10 |
ENSDART00000143325
|
nt5c1ba
|
5'-nucleotidase, cytosolic IB a |
| chr1_-_10473630 | 5.08 |
ENSDART00000040116
|
tnrc5
|
trinucleotide repeat containing 5 |
| chr9_+_38163876 | 5.04 |
ENSDART00000137955
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr20_-_19365875 | 4.96 |
ENSDART00000063703
ENSDART00000187707 ENSDART00000161065 |
si:dkey-71h2.2
|
si:dkey-71h2.2 |
| chr6_-_13408680 | 4.96 |
ENSDART00000151566
|
fmnl2b
|
formin-like 2b |
| chr4_+_6643421 | 4.95 |
ENSDART00000099462
|
gpr85
|
G protein-coupled receptor 85 |
| chr13_-_21701323 | 4.94 |
ENSDART00000164112
|
si:dkey-191g9.7
|
si:dkey-191g9.7 |
| chr11_+_18873619 | 4.93 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr24_-_6678640 | 4.84 |
ENSDART00000042478
|
enkur
|
enkurin, TRPC channel interacting protein |
| chr4_+_8168514 | 4.81 |
ENSDART00000150830
|
ninj2
|
ninjurin 2 |
| chr14_-_4682114 | 4.79 |
ENSDART00000014454
|
gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chr5_-_46896541 | 4.78 |
ENSDART00000133240
|
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr18_+_17428506 | 4.75 |
ENSDART00000100223
|
zgc:91860
|
zgc:91860 |
| chr16_-_17162843 | 4.75 |
ENSDART00000089386
|
iffo1b
|
intermediate filament family orphan 1b |
| chr16_-_52879741 | 4.64 |
ENSDART00000166470
|
TPPP
|
tubulin polymerization promoting protein |
| chr17_-_43391499 | 4.60 |
ENSDART00000189280
|
itpk1b
|
inositol-tetrakisphosphate 1-kinase b |
| chr5_+_52625975 | 4.59 |
ENSDART00000170341
ENSDART00000168317 |
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr16_-_37470042 | 4.59 |
ENSDART00000142916
|
si:ch211-208k15.1
|
si:ch211-208k15.1 |
| chr22_+_20195280 | 4.59 |
ENSDART00000088603
ENSDART00000135692 |
si:dkey-110c1.7
|
si:dkey-110c1.7 |
| chr17_+_31221761 | 4.56 |
ENSDART00000155580
|
ccdc32
|
coiled-coil domain containing 32 |
| chr18_+_7286788 | 4.53 |
ENSDART00000022998
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr19_+_24039830 | 4.53 |
ENSDART00000100422
|
rit1
|
Ras-like without CAAX 1 |
| chr9_+_29603649 | 4.53 |
ENSDART00000140477
|
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr19_+_16032383 | 4.50 |
ENSDART00000046530
|
rab42a
|
RAB42, member RAS oncogene family a |
| chr10_+_34426256 | 4.42 |
ENSDART00000102566
|
nbeaa
|
neurobeachin a |
| chr3_-_51912019 | 4.41 |
ENSDART00000149914
|
aatka
|
apoptosis-associated tyrosine kinase a |
| chr18_-_39787040 | 4.40 |
ENSDART00000169916
|
dmxl2
|
Dmx-like 2 |
| chr9_+_33154841 | 4.39 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr23_-_29505463 | 4.37 |
ENSDART00000050915
|
kif1b
|
kinesin family member 1B |
| chr12_-_4781801 | 4.37 |
ENSDART00000167490
ENSDART00000121718 |
mapta
|
microtubule-associated protein tau a |
| chr18_+_27337994 | 4.36 |
ENSDART00000136172
|
si:dkey-29p10.4
|
si:dkey-29p10.4 |
| chr3_-_26191960 | 4.35 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr23_+_31405497 | 4.32 |
ENSDART00000053546
|
sh3bgrl2
|
SH3 domain binding glutamate-rich protein like 2 |
| chr19_+_5604241 | 4.31 |
ENSDART00000011025
|
wipf2b
|
WAS/WASL interacting protein family, member 2b |
| chr5_-_40190949 | 4.31 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr10_+_22891126 | 4.27 |
ENSDART00000057291
|
arrb2a
|
arrestin, beta 2a |
| chr6_-_39605734 | 4.27 |
ENSDART00000044276
ENSDART00000179059 |
dip2bb
|
disco-interacting protein 2 homolog Bb |
| chr3_-_28258462 | 4.26 |
ENSDART00000191573
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr16_+_28578352 | 4.23 |
ENSDART00000149306
|
nmt2
|
N-myristoyltransferase 2 |
| chr13_-_22903246 | 4.21 |
ENSDART00000089133
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr2_-_11662851 | 4.19 |
ENSDART00000145108
|
zgc:110130
|
zgc:110130 |
| chr13_-_49169545 | 4.18 |
ENSDART00000192076
|
tsnax
|
translin-associated factor X |
| chr19_+_32979331 | 4.17 |
ENSDART00000078066
|
spire1a
|
spire-type actin nucleation factor 1a |
| chr15_+_8043751 | 4.17 |
ENSDART00000193701
|
cadm2b
|
cell adhesion molecule 2b |
| chr24_-_21989406 | 4.14 |
ENSDART00000032963
|
apoob
|
apolipoprotein O, b |
| chr7_+_52712807 | 4.11 |
ENSDART00000174095
ENSDART00000174377 ENSDART00000174061 ENSDART00000174094 ENSDART00000110906 ENSDART00000174071 ENSDART00000174238 |
znf280d
|
zinc finger protein 280D |
| chr11_-_24191928 | 4.10 |
ENSDART00000136827
|
sox12
|
SRY (sex determining region Y)-box 12 |
| chr20_+_42918755 | 4.10 |
ENSDART00000134855
|
efr3bb
|
EFR3 homolog Bb (S. cerevisiae) |
| chr20_-_26039841 | 4.07 |
ENSDART00000179929
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr5_-_46980651 | 4.07 |
ENSDART00000181022
ENSDART00000168038 |
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr24_-_22756508 | 4.03 |
ENSDART00000035409
ENSDART00000146247 |
zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr15_-_18574716 | 4.03 |
ENSDART00000142010
ENSDART00000019006 |
ncam1b
|
neural cell adhesion molecule 1b |
| chr20_+_40457599 | 4.01 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr13_-_12006007 | 3.94 |
ENSDART00000111438
|
mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr1_-_40994259 | 3.91 |
ENSDART00000101562
|
adra2c
|
adrenoceptor alpha 2C |
| chr2_-_42173834 | 3.91 |
ENSDART00000098357
ENSDART00000144707 |
slc39a6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr6_-_21616659 | 3.89 |
ENSDART00000074256
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr2_+_25929619 | 3.87 |
ENSDART00000137746
|
slc7a14a
|
solute carrier family 7, member 14a |
| chr20_+_30445971 | 3.86 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr11_+_6819050 | 3.86 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr13_-_11378355 | 3.85 |
ENSDART00000164566
|
akt3a
|
v-akt murine thymoma viral oncogene homolog 3a |
| chr2_-_3403020 | 3.85 |
ENSDART00000092741
|
snap47
|
synaptosomal-associated protein, 47 |
| chr3_+_32403758 | 3.83 |
ENSDART00000156982
|
si:ch211-195b15.8
|
si:ch211-195b15.8 |
| chr23_-_36449111 | 3.80 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr9_+_38962398 | 3.79 |
ENSDART00000134294
|
map2
|
microtubule-associated protein 2 |
| chr18_-_13121983 | 3.77 |
ENSDART00000092648
|
rxylt1
|
ribitol xylosyltransferase 1 |
| chr8_-_25716074 | 3.74 |
ENSDART00000007482
|
tspy
|
testis specific protein, Y-linked |
| chr14_-_4273396 | 3.72 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr5_+_51111343 | 3.70 |
ENSDART00000092002
|
pomt1
|
protein-O-mannosyltransferase 1 |
| chr23_+_19594608 | 3.69 |
ENSDART00000134865
|
slmapb
|
sarcolemma associated protein b |
| chr19_-_7358184 | 3.69 |
ENSDART00000092379
|
oxr1b
|
oxidation resistance 1b |
| chr5_+_15495351 | 3.67 |
ENSDART00000111646
ENSDART00000114446 |
suds3
|
SDS3 homolog, SIN3A corepressor complex component |
| chr11_+_29770966 | 3.67 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr23_-_15330168 | 3.66 |
ENSDART00000035865
ENSDART00000143635 |
sulf2b
|
sulfatase 2b |
| chr13_-_40499296 | 3.62 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr18_-_41219790 | 3.61 |
ENSDART00000162387
ENSDART00000193753 |
zbtb38
|
zinc finger and BTB domain containing 38 |
| chr25_+_10458990 | 3.61 |
ENSDART00000130354
ENSDART00000044738 |
ric8a
|
RIC8 guanine nucleotide exchange factor A |
| chr5_-_66749535 | 3.60 |
ENSDART00000132183
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr12_+_18578597 | 3.58 |
ENSDART00000134944
|
grid2ipb
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, b |
| chr14_+_4276394 | 3.54 |
ENSDART00000038301
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr9_+_19623363 | 3.53 |
ENSDART00000142471
ENSDART00000147662 ENSDART00000136053 |
pdxka
|
pyridoxal (pyridoxine, vitamin B6) kinase a |
| chr4_-_14207471 | 3.47 |
ENSDART00000015134
|
twf1b
|
twinfilin actin-binding protein 1b |
| chr13_-_12005429 | 3.47 |
ENSDART00000180302
|
mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr10_+_22890791 | 3.45 |
ENSDART00000176011
|
arrb2a
|
arrestin, beta 2a |
| chr9_-_3149896 | 3.45 |
ENSDART00000020861
|
pdk1
|
pyruvate dehydrogenase kinase, isozyme 1 |
| chr16_-_12173399 | 3.45 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr12_-_32066469 | 3.43 |
ENSDART00000140685
ENSDART00000062185 |
rab40b
|
RAB40B, member RAS oncogene family |
| chr23_-_30764319 | 3.40 |
ENSDART00000075918
|
pcmtd2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr13_-_18691041 | 3.40 |
ENSDART00000057867
|
sfxn3
|
sideroflexin 3 |
| chr1_+_21937201 | 3.39 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr12_-_7607114 | 3.37 |
ENSDART00000158095
|
slc16a9b
|
solute carrier family 16, member 9b |
| chr2_-_40135942 | 3.36 |
ENSDART00000176951
ENSDART00000098632 ENSDART00000148563 ENSDART00000149895 |
epha4a
|
eph receptor A4a |
| chr19_+_20178978 | 3.34 |
ENSDART00000145115
ENSDART00000151175 |
tra2a
|
transformer 2 alpha homolog |
| chr17_-_13026634 | 3.33 |
ENSDART00000113713
|
fam177a1
|
family with sequence similarity 177, member A1 |
| chr5_-_25236340 | 3.31 |
ENSDART00000162774
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr25_+_15647993 | 3.31 |
ENSDART00000186578
ENSDART00000031828 |
spon1b
|
spondin 1b |
| chr22_+_10713713 | 3.29 |
ENSDART00000122349
|
hiat1b
|
hippocampus abundant transcript 1b |
| chr20_+_29217835 | 3.29 |
ENSDART00000024528
|
emc7
|
ER membrane protein complex subunit 7 |
| chr5_+_34981584 | 3.28 |
ENSDART00000134795
|
ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr16_+_5612547 | 3.26 |
ENSDART00000140226
ENSDART00000189352 |
CYTH2
|
si:dkey-283b15.4 |
| chr11_+_30162407 | 3.25 |
ENSDART00000190333
ENSDART00000127502 |
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr7_-_31618166 | 3.24 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr24_-_20658446 | 3.23 |
ENSDART00000127923
|
nktr
|
natural killer cell triggering receptor |
| chr21_-_4032650 | 3.22 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr12_+_27213733 | 3.21 |
ENSDART00000133048
|
nbr1a
|
neighbor of brca1 gene 1a |
| chr7_+_22823889 | 3.21 |
ENSDART00000127467
ENSDART00000148576 ENSDART00000149993 |
pygmb
|
phosphorylase, glycogen, muscle b |
| chr5_-_28767573 | 3.19 |
ENSDART00000158299
ENSDART00000043466 |
traf2a
|
Tnf receptor-associated factor 2a |
| chr13_+_28612313 | 3.19 |
ENSDART00000077383
|
borcs7
|
BLOC-1 related complex subunit 7 |
| chr2_+_26240339 | 3.17 |
ENSDART00000191006
|
palm1b
|
paralemmin 1b |
| chr18_-_30020879 | 3.17 |
ENSDART00000162086
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr6_-_1553314 | 3.15 |
ENSDART00000077209
|
tpra1
|
transmembrane protein, adipocyte asscociated 1 |
| chr18_+_19419120 | 3.14 |
ENSDART00000025107
|
map2k1
|
mitogen-activated protein kinase kinase 1 |
| chr6_-_8392104 | 3.14 |
ENSDART00000081561
ENSDART00000181178 |
ilf3a
|
interleukin enhancer binding factor 3a |
| chr14_-_2352384 | 3.13 |
ENSDART00000170666
|
si:ch73-233f7.7
|
si:ch73-233f7.7 |
| chr13_+_22280983 | 3.12 |
ENSDART00000173258
ENSDART00000173379 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr25_+_388258 | 3.12 |
ENSDART00000166834
|
rfx7b
|
regulatory factor X7b |
| chr11_-_18601955 | 3.12 |
ENSDART00000180565
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr5_+_61799629 | 3.12 |
ENSDART00000113508
|
hnrnpul1l
|
heterogeneous nuclear ribonucleoprotein U-like 1 like |
| chr3_+_25907266 | 3.12 |
ENSDART00000170324
ENSDART00000192633 |
tom1
|
target of myb1 membrane trafficking protein |
| chr6_+_13933464 | 3.10 |
ENSDART00000109144
|
ptprnb
|
protein tyrosine phosphatase, receptor type, Nb |
| chr3_-_18792492 | 3.10 |
ENSDART00000134208
ENSDART00000034373 |
hagh
|
hydroxyacylglutathione hydrolase |
| chr17_+_20173882 | 3.08 |
ENSDART00000155379
|
si:ch211-248a14.8
|
si:ch211-248a14.8 |
| chr10_-_32880298 | 3.08 |
ENSDART00000138243
|
rabgef1
|
RAB guanine nucleotide exchange factor (GEF) 1 |
| chr7_+_48805534 | 3.05 |
ENSDART00000145375
ENSDART00000148744 |
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr2_+_38924975 | 3.04 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr19_-_19025998 | 3.04 |
ENSDART00000186156
ENSDART00000163359 ENSDART00000167951 |
dync1li1
|
dynein, cytoplasmic 1, light intermediate chain 1 |
| chr2_-_54054225 | 3.01 |
ENSDART00000167239
|
CABZ01050249.1
|
|
| chr21_+_8427059 | 2.99 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr4_-_4261673 | 2.98 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxb1b+foxb2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.8 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 2.3 | 20.8 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 2.1 | 6.4 | GO:1903332 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 2.0 | 6.1 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 1.9 | 5.8 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 1.9 | 11.6 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 1.9 | 7.7 | GO:1900136 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 1.7 | 5.1 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 1.6 | 6.6 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 1.6 | 6.4 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 1.5 | 4.6 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 1.5 | 6.1 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 1.5 | 4.4 | GO:0097401 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 1.3 | 3.9 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 1.2 | 5.9 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 1.2 | 3.5 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 1.2 | 3.5 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 1.1 | 4.6 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 1.1 | 1.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 1.1 | 3.2 | GO:0052575 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 1.0 | 3.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 1.0 | 15.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 1.0 | 6.9 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 1.0 | 8.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 1.0 | 2.9 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 1.0 | 2.9 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.9 | 6.5 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.9 | 6.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.8 | 2.5 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.8 | 5.8 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.8 | 4.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.8 | 3.9 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.8 | 4.6 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.7 | 4.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.7 | 11.6 | GO:0035268 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.7 | 3.5 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.7 | 2.7 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.7 | 3.4 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.6 | 1.9 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.6 | 3.0 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.6 | 1.8 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.6 | 1.8 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.6 | 2.8 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 0.5 | 2.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.5 | 5.4 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.5 | 8.0 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.5 | 2.7 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.5 | 4.2 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.5 | 6.3 | GO:0001964 | startle response(GO:0001964) |
| 0.5 | 5.4 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.5 | 8.5 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.5 | 8.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.5 | 9.4 | GO:0007032 | endosome organization(GO:0007032) |
| 0.5 | 2.8 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.5 | 1.8 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.4 | 2.7 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.4 | 3.9 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.4 | 6.0 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.4 | 2.6 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.4 | 6.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.4 | 1.2 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.4 | 0.4 | GO:0061195 | tongue development(GO:0043586) tongue morphogenesis(GO:0043587) taste bud development(GO:0061193) taste bud morphogenesis(GO:0061194) taste bud formation(GO:0061195) |
| 0.4 | 2.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.4 | 2.8 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.3 | 9.8 | GO:0051923 | sulfation(GO:0051923) |
| 0.3 | 2.0 | GO:1900044 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.3 | 3.7 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.3 | 6.3 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.3 | 2.3 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.3 | 5.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.3 | 3.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.3 | 2.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.3 | 3.0 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.3 | 8.2 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.3 | 1.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.3 | 1.5 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.3 | 1.5 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.3 | 1.5 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.3 | 1.8 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.3 | 1.8 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.3 | 1.7 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.3 | 0.9 | GO:1904871 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.3 | 2.0 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.3 | 2.0 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.3 | 1.4 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.3 | 12.4 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.3 | 4.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.3 | 1.9 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.3 | 3.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.3 | 2.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.3 | 3.7 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.3 | 1.6 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.3 | 0.8 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.3 | 1.3 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.2 | 2.0 | GO:0060118 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.2 | 2.5 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.2 | 0.7 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 1.2 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.2 | 0.9 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.2 | 1.4 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.2 | 1.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 5.5 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.2 | 1.8 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 | 4.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 1.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 0.9 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.2 | 2.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 1.6 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.2 | 6.7 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.2 | 1.3 | GO:0090660 | cerebrospinal fluid circulation(GO:0090660) |
| 0.2 | 0.9 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.2 | 0.9 | GO:0050955 | thermoception(GO:0050955) detection of temperature stimulus involved in thermoception(GO:0050960) detection of temperature stimulus involved in sensory perception(GO:0050961) |
| 0.2 | 9.9 | GO:1902749 | regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.2 | 4.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.2 | 5.1 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.2 | 0.9 | GO:0042779 | tRNA 3'-trailer cleavage(GO:0042779) |
| 0.2 | 2.9 | GO:0030317 | sperm motility(GO:0030317) |
| 0.2 | 4.8 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.2 | 1.6 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.2 | 3.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.2 | 0.6 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.2 | 1.4 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.2 | 1.6 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 5.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.2 | 2.7 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.2 | 3.2 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.2 | 1.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.2 | 1.1 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.2 | 3.5 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.2 | 1.3 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 0.7 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.2 | 0.7 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.2 | 1.7 | GO:0034340 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.2 | 2.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 0.8 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.2 | 4.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.2 | 4.2 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.2 | 7.1 | GO:0030901 | midbrain development(GO:0030901) |
| 0.2 | 1.1 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.2 | 0.6 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 | 6.3 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.2 | 5.0 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.2 | 2.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 1.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 2.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 2.0 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.1 | 1.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 1.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 2.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.4 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 2.7 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 2.4 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.8 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 2.0 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 3.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.6 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 2.1 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 2.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 1.5 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.1 | 3.8 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 6.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 0.9 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 4.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 1.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.8 | GO:0007612 | learning(GO:0007612) nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 0.7 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 3.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.4 | GO:0018008 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.1 | 0.7 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 6.2 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 10.1 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.1 | 0.4 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 2.0 | GO:0048488 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 0.4 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.1 | 2.0 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 1.2 | GO:0021986 | habenula development(GO:0021986) |
| 0.1 | 2.4 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.1 | 2.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 1.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 2.3 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 2.6 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 4.0 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.1 | 1.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 7.8 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 4.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 2.3 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 2.2 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 1.2 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 0.3 | GO:0006750 | glutathione biosynthetic process(GO:0006750) nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.1 | 0.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 7.5 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.1 | 4.3 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.1 | 2.5 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.1 | 1.3 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.1 | 2.2 | GO:0007031 | peroxisome organization(GO:0007031) |
| 0.1 | 1.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.9 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.1 | 1.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 1.3 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.1 | 2.2 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.2 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.5 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 1.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.9 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 0.6 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.1 | 1.6 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 4.7 | GO:0009615 | response to virus(GO:0009615) |
| 0.1 | 0.2 | GO:0051928 | positive regulation of calcium ion transport(GO:0051928) |
| 0.1 | 3.5 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 11.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.7 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 1.8 | GO:0010906 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 1.5 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.2 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 20.3 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 2.6 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.0 | 0.7 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.0 | 1.9 | GO:0022406 | membrane docking(GO:0022406) vesicle docking(GO:0048278) |
| 0.0 | 0.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 1.4 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 3.7 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 2.7 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 1.9 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 3.4 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 3.6 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 1.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.5 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 3.5 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 1.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.2 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 8.7 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 2.4 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.3 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.7 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.1 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 2.2 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 14.2 | GO:0031175 | neuron projection development(GO:0031175) |
| 0.0 | 2.7 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 1.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.3 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.0 | 0.3 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.0 | 0.1 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 5.5 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.5 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 2.1 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 1.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0016116 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.5 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 7.8 | GO:0098662 | inorganic cation transmembrane transport(GO:0098662) |
| 0.0 | 1.0 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.5 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 1.6 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.5 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 4.2 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 2.6 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.4 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.8 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 15.5 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 2.1 | 6.4 | GO:0072380 | TRC complex(GO:0072380) |
| 1.7 | 5.0 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 1.6 | 9.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.5 | 6.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 1.1 | 4.6 | GO:0035339 | SPOTS complex(GO:0035339) |
| 1.1 | 4.4 | GO:0043291 | RAVE complex(GO:0043291) |
| 1.1 | 3.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 1.0 | 5.9 | GO:0070695 | FHF complex(GO:0070695) |
| 0.8 | 2.5 | GO:0031213 | RSF complex(GO:0031213) |
| 0.6 | 6.7 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.6 | 8.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.5 | 4.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.5 | 3.8 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.4 | 1.7 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.4 | 2.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.4 | 1.6 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.4 | 6.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 4.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.3 | 1.4 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.3 | 2.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.3 | 5.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 2.7 | GO:0016586 | RSC complex(GO:0016586) |
| 0.3 | 9.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.3 | 2.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.3 | 1.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.3 | 2.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.3 | 1.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 1.5 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.3 | 7.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 2.8 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.3 | 11.8 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.3 | 2.2 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.3 | 1.9 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.3 | 1.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 0.9 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.2 | 3.4 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.2 | 2.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 3.0 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 0.6 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 1.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 4.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.2 | 6.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 1.0 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 4.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 16.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 6.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.2 | 2.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 3.9 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.2 | 5.1 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.2 | 4.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.2 | 6.0 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.2 | 0.9 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 1.7 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.2 | 1.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 4.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 1.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 25.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 1.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.4 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 3.0 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 5.3 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 0.7 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 1.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 16.0 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.1 | 8.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 2.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 5.9 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 4.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 3.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 2.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 1.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 5.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.9 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.9 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.6 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 1.3 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.1 | 26.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 1.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.9 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 11.8 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 16.0 | GO:0098791 | Golgi subcompartment(GO:0098791) |
| 0.1 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 2.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.4 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 7.9 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 3.4 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 8.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 1.8 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 3.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 3.3 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 10.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 2.0 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 0.8 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 2.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 1.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 2.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 5.9 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 2.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 27.1 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 3.8 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.5 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 3.9 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 5.2 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 2.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 25.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.3 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 2.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 4.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.9 | GO:0005635 | nuclear envelope(GO:0005635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.7 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 1.6 | 6.6 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 1.6 | 9.3 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 1.5 | 4.6 | GO:0047325 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 1.5 | 6.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 1.4 | 5.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 1.2 | 15.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.2 | 3.5 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.9 | 2.7 | GO:0051765 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol tetrakisphosphate kinase activity(GO:0051765) inositol trisphosphate kinase activity(GO:0051766) |
| 0.9 | 8.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.9 | 7.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.8 | 5.8 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.8 | 2.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.7 | 2.2 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.7 | 2.7 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.7 | 11.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.7 | 2.7 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.7 | 2.7 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.7 | 19.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.7 | 3.9 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.6 | 3.2 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.6 | 8.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.6 | 1.8 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.6 | 3.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.6 | 3.4 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.6 | 2.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.6 | 7.9 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.6 | 5.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.5 | 8.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.5 | 6.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.5 | 1.9 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.5 | 1.9 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.5 | 1.8 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.4 | 14.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.4 | 1.8 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.4 | 3.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.4 | 3.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.4 | 2.1 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.4 | 1.2 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.4 | 2.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.4 | 9.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.4 | 5.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.4 | 1.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.4 | 6.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.4 | 2.8 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.3 | 4.8 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.3 | 2.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 2.0 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.3 | 3.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.3 | 1.6 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.3 | 6.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.3 | 2.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.3 | 1.5 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.3 | 2.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.3 | 3.8 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.3 | 1.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.3 | 0.8 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.3 | 3.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 2.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 4.9 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.3 | 1.3 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.3 | 2.5 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.2 | 2.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 1.0 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.2 | 2.0 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.2 | 0.7 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 0.9 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 1.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 5.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 2.7 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 3.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 5.5 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.2 | 2.7 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 1.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 1.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 2.9 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.2 | 7.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 3.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.2 | 3.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 0.6 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 3.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 5.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 0.8 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.2 | 7.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.2 | 2.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 3.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.2 | 0.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 3.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.6 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.4 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 2.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 3.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 2.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 2.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 48.7 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 3.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 2.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.9 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.4 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.1 | 9.8 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 5.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 9.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 6.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.4 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.1 | 16.9 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.1 | 1.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.5 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 1.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 2.2 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 0.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 3.1 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 0.2 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.1 | 0.5 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.5 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 0.5 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 3.6 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 1.9 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 4.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 14.1 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 2.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 3.5 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 1.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 4.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.4 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.5 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 1.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.1 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 6.2 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 3.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 2.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 2.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 15.5 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.6 | GO:0008443 | 6-phosphofructo-2-kinase activity(GO:0003873) phosphofructokinase activity(GO:0008443) |
| 0.0 | 4.1 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 21.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.7 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.9 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 3.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 1.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.1 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 1.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 8.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 5.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.7 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 5.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 2.3 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.9 | GO:0015278 | calcium-release channel activity(GO:0015278) |
| 0.0 | 1.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.2 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.9 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 1.6 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.1 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.0 | 7.5 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 2.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.6 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 1.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.5 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 1.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.8 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 10.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.5 | GO:0045296 | cadherin binding(GO:0045296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.4 | 2.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.3 | 3.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 1.7 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.3 | 12.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 5.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 2.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 3.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 6.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 2.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 4.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 4.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 4.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 2.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 5.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 1.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 6.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 3.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.7 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 2.3 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 3.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 1.5 | 15.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.9 | 7.7 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.5 | 9.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.5 | 3.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.5 | 2.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.4 | 4.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.3 | 2.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 4.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.3 | 3.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 2.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.3 | 3.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 3.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 4.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 14.4 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.2 | 2.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 0.6 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.2 | 1.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 1.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 5.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 1.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 1.6 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.2 | 2.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 2.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 2.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 2.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 1.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.8 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 13.0 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 1.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 0.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.0 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.8 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 2.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.1 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |