Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
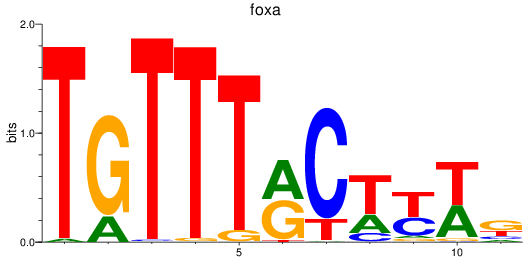
Results for foxa_foxa1_foxa2
Z-value: 2.57
Transcription factors associated with foxa_foxa1_foxa2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxa
|
ENSDARG00000087094 | forkhead box A sequence |
|
foxa
|
ENSDARG00000110743 | forkhead box A sequence |
|
foxa
|
ENSDARG00000115019 | forkhead box A sequence |
|
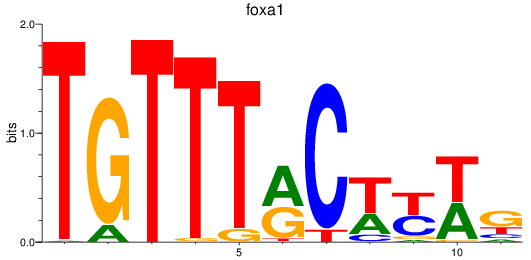
foxa1
|
ENSDARG00000102138 | forkhead box A1 |
|
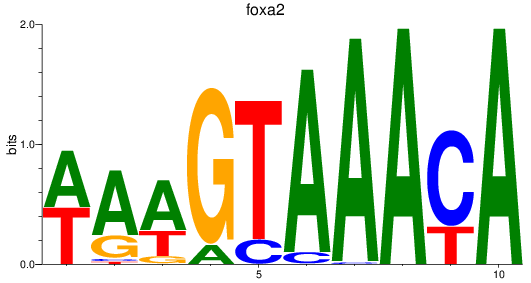
foxa2
|
ENSDARG00000003411 | forkhead box A2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxa2 | dr11_v1_chr17_-_42568498_42568498 | 0.84 | 3.5e-26 | Click! |
| foxa1 | dr11_v1_chr17_+_10318071_10318071 | 0.58 | 1.2e-09 | Click! |
| foxa | dr11_v1_chr14_-_33981544_33981544 | 0.08 | 4.3e-01 | Click! |
Activity profile of foxa_foxa1_foxa2 motif
Sorted Z-values of foxa_foxa1_foxa2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_6142433 | 84.64 |
ENSDART00000054084
ENSDART00000136986 |
ttr
|
transthyretin (prealbumin, amyloidosis type I) |
| chr5_+_28830643 | 68.08 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr16_+_23978978 | 68.02 |
ENSDART00000058964
ENSDART00000135084 |
apoa2
|
apolipoprotein A-II |
| chr22_-_36856405 | 66.57 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr5_+_28830388 | 63.86 |
ENSDART00000149150
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr2_+_10134345 | 63.40 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr5_+_28857969 | 62.21 |
ENSDART00000149850
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr16_-_54455573 | 61.37 |
ENSDART00000075275
|
pklr
|
pyruvate kinase L/R |
| chr5_+_28848870 | 59.80 |
ENSDART00000149563
|
zgc:174259
|
zgc:174259 |
| chr5_+_28849155 | 58.15 |
ENSDART00000079090
|
zgc:174259
|
zgc:174259 |
| chr23_+_28770225 | 57.63 |
ENSDART00000132179
ENSDART00000142273 |
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr5_+_28797771 | 56.55 |
ENSDART00000188845
ENSDART00000149066 |
si:ch211-186e20.7
|
si:ch211-186e20.7 |
| chr5_+_28858345 | 45.89 |
ENSDART00000111180
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr2_+_16781015 | 43.46 |
ENSDART00000155147
ENSDART00000003845 |
tfa
|
transferrin-a |
| chr2_+_16780643 | 42.49 |
ENSDART00000125647
ENSDART00000108611 ENSDART00000181245 ENSDART00000163194 |
tfa
|
transferrin-a |
| chr6_-_55864687 | 41.34 |
ENSDART00000160991
|
cyp24a1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr15_+_14856307 | 40.70 |
ENSDART00000167213
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr10_+_17026870 | 37.76 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr4_-_16824556 | 37.13 |
ENSDART00000165289
ENSDART00000185839 |
gys2
|
glycogen synthase 2 |
| chr4_-_16824231 | 36.77 |
ENSDART00000014007
|
gys2
|
glycogen synthase 2 |
| chr12_+_31616412 | 35.69 |
ENSDART00000124439
|
cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr10_+_26747755 | 30.73 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr22_-_24967348 | 29.42 |
ENSDART00000153490
ENSDART00000084871 |
fam20cl
|
family with sequence similarity 20, member C like |
| chr16_-_31661536 | 28.97 |
ENSDART00000169973
|
wu:fd46c06
|
wu:fd46c06 |
| chr10_+_7709724 | 28.93 |
ENSDART00000097670
|
ggcx
|
gamma-glutamyl carboxylase |
| chr23_+_44881020 | 28.03 |
ENSDART00000149355
|
si:ch73-361h17.1
|
si:ch73-361h17.1 |
| chr20_+_23440632 | 27.64 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr14_-_36799280 | 26.63 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr23_-_31763753 | 25.34 |
ENSDART00000053399
|
aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr6_+_6780873 | 22.85 |
ENSDART00000011865
|
sec23b
|
Sec23 homolog B, COPII coat complex component |
| chr4_+_1530287 | 21.18 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
| chr17_+_24036791 | 20.99 |
ENSDART00000140767
|
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr21_+_27416284 | 20.58 |
ENSDART00000077593
ENSDART00000108763 |
cfb
|
complement factor B |
| chr4_+_18806251 | 19.46 |
ENSDART00000138662
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr7_+_69019851 | 19.25 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr20_+_26881600 | 18.04 |
ENSDART00000174799
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr2_+_1486822 | 17.94 |
ENSDART00000132500
|
c8a
|
complement component 8, alpha polypeptide |
| chr2_+_1487118 | 17.81 |
ENSDART00000147283
|
c8a
|
complement component 8, alpha polypeptide |
| chr13_+_23988442 | 17.04 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr16_+_14588141 | 17.02 |
ENSDART00000140469
ENSDART00000059984 ENSDART00000167411 ENSDART00000133566 |
deptor
|
DEP domain containing MTOR-interacting protein |
| chr8_+_30709685 | 16.96 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr5_+_65991152 | 16.69 |
ENSDART00000097756
|
lcn15
|
lipocalin 15 |
| chr24_+_26887487 | 16.62 |
ENSDART00000189425
|
CR376848.1
|
|
| chr4_+_2230701 | 16.34 |
ENSDART00000080439
|
cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr18_-_6803424 | 15.85 |
ENSDART00000142647
|
si:dkey-266m15.5
|
si:dkey-266m15.5 |
| chr21_+_40092301 | 15.83 |
ENSDART00000145150
|
serpinf2a
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2a |
| chr2_-_38035235 | 15.56 |
ENSDART00000075904
|
cbln5
|
cerebellin 5 |
| chr3_+_46762703 | 15.49 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr3_-_55525627 | 15.28 |
ENSDART00000189234
|
tex2
|
testis expressed 2 |
| chr14_+_15155684 | 14.93 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr23_+_39606108 | 14.71 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr24_+_22056386 | 14.57 |
ENSDART00000132390
|
ankrd33ba
|
ankyrin repeat domain 33ba |
| chr11_-_3629201 | 14.46 |
ENSDART00000136577
ENSDART00000132121 |
itih3a
|
inter-alpha-trypsin inhibitor heavy chain 3a |
| chr5_+_32345187 | 14.44 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr1_-_25177086 | 14.31 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr23_-_33750307 | 14.17 |
ENSDART00000162772
|
bin2a
|
bridging integrator 2a |
| chr5_-_66160415 | 14.16 |
ENSDART00000073895
|
mboat4
|
membrane bound O-acyltransferase domain containing 4 |
| chr15_+_36187434 | 13.80 |
ENSDART00000181536
ENSDART00000099501 ENSDART00000154432 |
masp1
|
mannan-binding lectin serine peptidase 1 |
| chr23_-_33750135 | 13.77 |
ENSDART00000187641
|
bin2a
|
bridging integrator 2a |
| chr25_-_13188678 | 13.63 |
ENSDART00000125754
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr8_+_49570884 | 13.41 |
ENSDART00000182117
ENSDART00000108613 |
rasef
|
RAS and EF-hand domain containing |
| chr20_-_36809059 | 13.39 |
ENSDART00000062925
|
slc25a27
|
solute carrier family 25, member 27 |
| chr19_+_7636941 | 13.35 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr16_-_21785261 | 13.35 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr16_-_17713859 | 13.30 |
ENSDART00000149275
|
zgc:174935
|
zgc:174935 |
| chr21_-_39081107 | 13.26 |
ENSDART00000075935
|
vtnb
|
vitronectin b |
| chr2_-_43168292 | 13.18 |
ENSDART00000132588
|
crema
|
cAMP responsive element modulator a |
| chr18_-_14777092 | 13.18 |
ENSDART00000144660
|
mtss1la
|
metastasis suppressor 1-like a |
| chr4_+_17310143 | 13.17 |
ENSDART00000004717
|
igf1
|
insulin-like growth factor 1 |
| chr20_-_19365875 | 12.87 |
ENSDART00000063703
ENSDART00000187707 ENSDART00000161065 |
si:dkey-71h2.2
|
si:dkey-71h2.2 |
| chr24_-_26369185 | 12.75 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr5_+_45677781 | 12.75 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr23_-_25686894 | 12.63 |
ENSDART00000181420
ENSDART00000088208 |
lrp1ab
|
low density lipoprotein receptor-related protein 1Ab |
| chr13_-_8692860 | 12.62 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr2_+_30969029 | 12.32 |
ENSDART00000085242
|
lpin2
|
lipin 2 |
| chr14_+_32918484 | 12.30 |
ENSDART00000105721
|
lnx2b
|
ligand of numb-protein X 2b |
| chr7_+_48805534 | 12.15 |
ENSDART00000145375
ENSDART00000148744 |
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr11_+_41936435 | 12.13 |
ENSDART00000173103
|
aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr7_+_48805725 | 11.91 |
ENSDART00000166543
|
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr9_-_38036984 | 11.68 |
ENSDART00000134574
|
hacd2
|
3-hydroxyacyl-CoA dehydratase 2 |
| chr13_-_40726865 | 11.46 |
ENSDART00000099847
|
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr13_-_8692432 | 11.29 |
ENSDART00000058106
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr12_-_22238004 | 11.29 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr22_-_26236188 | 11.28 |
ENSDART00000162640
ENSDART00000167169 ENSDART00000138595 |
c3b.1
|
complement component c3b, tandem duplicate 1 |
| chr3_+_19299309 | 11.24 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr11_-_4235811 | 10.96 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr24_-_29963858 | 10.78 |
ENSDART00000183442
|
CR352310.1
|
|
| chr22_+_24559947 | 10.74 |
ENSDART00000169847
|
wdr47b
|
WD repeat domain 47b |
| chr5_-_31773208 | 10.71 |
ENSDART00000137556
ENSDART00000122066 |
fam102ab
|
family with sequence similarity 102, member Ab |
| chr11_+_13223625 | 10.08 |
ENSDART00000161275
|
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr11_+_25430851 | 9.91 |
ENSDART00000164999
ENSDART00000126403 |
si:dkey-13a21.4
|
si:dkey-13a21.4 |
| chr4_-_17725008 | 9.82 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr17_-_26867725 | 9.73 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr24_-_31306724 | 9.71 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr4_+_5868034 | 9.61 |
ENSDART00000166591
|
utp20
|
UTP20 small subunit (SSU) processome component |
| chr11_-_15296805 | 9.53 |
ENSDART00000124968
|
rpn2
|
ribophorin II |
| chr19_-_46091497 | 9.53 |
ENSDART00000178772
ENSDART00000167255 |
ptdss1b
si:dkey-108k24.2
|
phosphatidylserine synthase 1b si:dkey-108k24.2 |
| chr18_+_17611627 | 9.53 |
ENSDART00000046891
|
cetp
|
cholesteryl ester transfer protein, plasma |
| chr24_-_25461267 | 9.43 |
ENSDART00000105820
|
mbtps2
|
membrane-bound transcription factor peptidase, site 2 |
| chr2_-_5475910 | 9.32 |
ENSDART00000100954
ENSDART00000172143 ENSDART00000132496 |
proca
proca
|
protein C (inactivator of coagulation factors Va and VIIIa), a protein C (inactivator of coagulation factors Va and VIIIa), a |
| chr7_-_31759394 | 9.18 |
ENSDART00000193040
|
igdcc4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr2_+_31671545 | 9.04 |
ENSDART00000145446
|
ackr4a
|
atypical chemokine receptor 4a |
| chr16_-_25606235 | 8.78 |
ENSDART00000192741
|
zgc:110410
|
zgc:110410 |
| chr20_+_6590220 | 8.76 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr2_+_11028923 | 8.74 |
ENSDART00000076725
|
acot11a
|
acyl-CoA thioesterase 11a |
| chr4_-_8040436 | 8.72 |
ENSDART00000113033
|
si:ch211-240l19.6
|
si:ch211-240l19.6 |
| chr5_-_31772559 | 8.65 |
ENSDART00000183879
|
fam102ab
|
family with sequence similarity 102, member Ab |
| chr10_-_5135788 | 8.41 |
ENSDART00000108587
ENSDART00000138537 |
sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr11_+_18612421 | 8.23 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr5_-_34875858 | 8.21 |
ENSDART00000085086
|
arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr14_-_11529311 | 8.21 |
ENSDART00000127208
|
si:ch211-153b23.7
|
si:ch211-153b23.7 |
| chr10_+_11260170 | 8.20 |
ENSDART00000155742
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr9_+_33154841 | 8.13 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr21_-_11856143 | 8.12 |
ENSDART00000151204
|
ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr24_+_37723362 | 8.07 |
ENSDART00000136836
|
rab11fip3
|
RAB11 family interacting protein 3 (class II) |
| chr15_-_28247583 | 8.06 |
ENSDART00000112967
|
rilp
|
Rab interacting lysosomal protein |
| chr19_-_42588510 | 7.82 |
ENSDART00000102583
|
sytl1
|
synaptotagmin-like 1 |
| chr21_-_26028205 | 7.76 |
ENSDART00000034875
|
sdf2
|
stromal cell-derived factor 2 |
| chr13_-_33207367 | 7.75 |
ENSDART00000146138
ENSDART00000109667 ENSDART00000182741 |
trip11
|
thyroid hormone receptor interactor 11 |
| chr19_-_7272921 | 7.75 |
ENSDART00000102075
ENSDART00000132887 ENSDART00000130234 ENSDART00000193535 ENSDART00000136528 |
rxrba
|
retinoid x receptor, beta a |
| chr2_-_5466708 | 7.70 |
ENSDART00000136682
|
proca
|
protein C (inactivator of coagulation factors Va and VIIIa), a |
| chr15_-_25365319 | 7.68 |
ENSDART00000152651
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr23_-_10722664 | 7.61 |
ENSDART00000146526
ENSDART00000129022 ENSDART00000104985 |
foxp1a
|
forkhead box P1a |
| chr18_-_18543358 | 7.55 |
ENSDART00000126460
|
il34
|
interleukin 34 |
| chr6_-_9565526 | 7.37 |
ENSDART00000151470
|
map3k2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr16_-_25606889 | 7.35 |
ENSDART00000077447
ENSDART00000131528 |
zgc:110410
|
zgc:110410 |
| chr18_-_40901707 | 7.28 |
ENSDART00000139352
|
foxg1c
|
forkhead box G1c |
| chr24_-_2843107 | 7.26 |
ENSDART00000165290
|
cyb5a
|
cytochrome b5 type A (microsomal) |
| chr23_+_24085531 | 7.02 |
ENSDART00000139710
|
ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr14_-_30390145 | 6.96 |
ENSDART00000045423
|
slc7a2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr2_+_20539402 | 6.96 |
ENSDART00000129585
|
si:ch73-14h1.2
|
si:ch73-14h1.2 |
| chr10_-_3416258 | 6.92 |
ENSDART00000005168
|
ddx55
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 |
| chr10_+_41199660 | 6.92 |
ENSDART00000125314
|
adrb3b
|
adrenoceptor beta 3b |
| chr7_+_9904627 | 6.71 |
ENSDART00000172824
|
cers3a
|
ceramide synthase 3a |
| chr22_-_17595310 | 6.67 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr17_-_30652738 | 6.66 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr18_-_44847855 | 6.50 |
ENSDART00000086823
|
srpr
|
signal recognition particle receptor (docking protein) |
| chr8_-_25605537 | 6.47 |
ENSDART00000005906
|
stk38a
|
serine/threonine kinase 38a |
| chr12_-_1361517 | 6.45 |
ENSDART00000188297
|
LO018020.2
|
|
| chr21_-_11855828 | 6.42 |
ENSDART00000081666
|
ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr17_-_23709347 | 6.41 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr12_-_28537615 | 6.40 |
ENSDART00000067762
|
MYO1D
|
si:ch211-94l19.4 |
| chr25_-_12805295 | 6.30 |
ENSDART00000157629
|
ca5a
|
carbonic anhydrase Va |
| chr18_+_40354998 | 6.22 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr16_+_52105227 | 6.14 |
ENSDART00000150025
ENSDART00000097863 |
MAN1C1
|
si:ch73-373m9.1 |
| chr14_+_32926385 | 6.11 |
ENSDART00000139159
|
lnx2b
|
ligand of numb-protein X 2b |
| chr11_-_11336986 | 6.08 |
ENSDART00000016677
|
zgc:77929
|
zgc:77929 |
| chr16_-_34477805 | 6.08 |
ENSDART00000136546
|
serinc2l
|
serine incorporator 2, like |
| chr4_+_5848229 | 6.07 |
ENSDART00000161101
ENSDART00000067357 |
lyrm5a
|
LYR motif containing 5a |
| chr11_+_44503774 | 5.95 |
ENSDART00000169295
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr5_+_34622320 | 5.90 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr11_+_35171406 | 5.89 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr21_-_22398985 | 5.88 |
ENSDART00000138128
|
si:ch73-112l6.1
|
si:ch73-112l6.1 |
| chr5_-_16351306 | 5.82 |
ENSDART00000168643
|
CABZ01088700.1
|
|
| chr16_-_25608453 | 5.68 |
ENSDART00000140140
|
zgc:110410
|
zgc:110410 |
| chr13_+_18545819 | 5.68 |
ENSDART00000101859
ENSDART00000110197 ENSDART00000136064 |
zgc:154058
|
zgc:154058 |
| chr25_-_13188214 | 5.66 |
ENSDART00000187298
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr15_-_30816370 | 5.54 |
ENSDART00000142982
ENSDART00000050649 ENSDART00000136901 ENSDART00000100194 |
msi2b
|
musashi RNA-binding protein 2b |
| chr23_-_10723009 | 5.26 |
ENSDART00000189721
|
foxp1a
|
forkhead box P1a |
| chr24_+_7782313 | 5.15 |
ENSDART00000111090
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr8_+_24747865 | 5.14 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr20_-_52928541 | 5.03 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr18_-_24988645 | 4.99 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr20_-_10288156 | 4.97 |
ENSDART00000064110
|
si:dkey-63b1.1
|
si:dkey-63b1.1 |
| chr10_+_11261576 | 4.97 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr18_-_6855991 | 4.97 |
ENSDART00000135206
|
ppp6r2b
|
protein phosphatase 6, regulatory subunit 2b |
| chr13_-_18345854 | 4.90 |
ENSDART00000080107
|
si:dkey-228d14.5
|
si:dkey-228d14.5 |
| chr17_+_32158951 | 4.89 |
ENSDART00000165348
ENSDART00000108736 |
adam12
|
ADAM metallopeptidase domain 12 |
| chr17_+_32623931 | 4.86 |
ENSDART00000144217
|
ctsba
|
cathepsin Ba |
| chr20_-_25902141 | 4.84 |
ENSDART00000142611
ENSDART00000024821 |
elmsan1a
|
ELM2 and Myb/SANT-like domain containing 1a |
| chr16_+_19637384 | 4.81 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr16_+_19029297 | 4.78 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr8_-_29822527 | 4.70 |
ENSDART00000167487
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr9_-_29497916 | 4.68 |
ENSDART00000060246
|
dnajc3a
|
DnaJ (Hsp40) homolog, subfamily C, member 3a |
| chr7_-_28549361 | 4.65 |
ENSDART00000173918
ENSDART00000054368 ENSDART00000113313 |
st5
|
suppression of tumorigenicity 5 |
| chr21_-_20840714 | 4.63 |
ENSDART00000144861
ENSDART00000139430 |
c6
|
complement component 6 |
| chr15_-_25365570 | 4.61 |
ENSDART00000152754
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr22_-_24285432 | 4.59 |
ENSDART00000164083
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr22_+_105376 | 4.53 |
ENSDART00000059140
|
slc25a20
|
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chr15_-_23793641 | 4.31 |
ENSDART00000122891
|
tmem97
|
transmembrane protein 97 |
| chr7_+_61184551 | 4.30 |
ENSDART00000190788
|
zgc:194930
|
zgc:194930 |
| chr7_+_9308625 | 4.28 |
ENSDART00000084598
|
selenos
|
selenoprotein S |
| chr3_-_55537096 | 4.26 |
ENSDART00000123544
ENSDART00000188752 |
tex2
|
testis expressed 2 |
| chr6_-_43677125 | 4.20 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr17_+_25519089 | 4.17 |
ENSDART00000041721
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr1_-_31856622 | 4.07 |
ENSDART00000065125
|
nt5c2b
|
5'-nucleotidase, cytosolic IIb |
| chr2_-_42492201 | 4.07 |
ENSDART00000180762
ENSDART00000009093 |
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr8_-_1219815 | 4.06 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr16_-_21038015 | 4.03 |
ENSDART00000059239
|
snx10b
|
sorting nexin 10b |
| chr2_-_29994726 | 3.99 |
ENSDART00000163350
|
cnpy1
|
canopy1 |
| chr15_-_8191841 | 3.80 |
ENSDART00000156663
|
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr15_+_38299563 | 3.76 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr21_+_26071874 | 3.74 |
ENSDART00000003001
ENSDART00000146573 |
rpl23a
|
ribosomal protein L23a |
| chr3_-_56871330 | 3.72 |
ENSDART00000014103
|
zgc:112148
|
zgc:112148 |
| chr7_-_71384391 | 3.64 |
ENSDART00000112841
|
ccdc149a
|
coiled-coil domain containing 149a |
| chr14_-_28052474 | 3.59 |
ENSDART00000172948
ENSDART00000135337 |
TSC22D3 (1 of many)
zgc:64189
|
si:ch211-220e11.3 zgc:64189 |
| chr7_+_58730201 | 3.58 |
ENSDART00000073640
|
plag1
|
pleiomorphic adenoma gene 1 |
| chr8_-_25771474 | 3.55 |
ENSDART00000193883
|
suv39h1b
|
suppressor of variegation 3-9 homolog 1b |
| chr17_+_33415319 | 3.49 |
ENSDART00000140805
ENSDART00000025501 ENSDART00000146447 |
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr10_-_13343831 | 3.46 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxa_foxa1_foxa2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.8 | 41.3 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 9.8 | 68.6 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 8.5 | 84.6 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 8.4 | 25.3 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 8.1 | 40.7 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 7.8 | 85.9 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 7.4 | 29.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 6.2 | 524.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 4.8 | 28.9 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 4.8 | 14.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 4.7 | 27.9 | GO:0071800 | podosome assembly(GO:0071800) |
| 3.6 | 57.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 3.4 | 17.0 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 3.2 | 77.9 | GO:0009749 | response to glucose(GO:0009749) |
| 3.2 | 9.5 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 2.8 | 11.3 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 2.5 | 10.1 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 2.5 | 7.6 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 2.4 | 12.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 2.4 | 24.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 2.1 | 6.4 | GO:0000103 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 2.1 | 31.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 1.9 | 9.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 1.9 | 11.2 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 1.8 | 10.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 1.7 | 3.3 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 1.6 | 16.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 1.5 | 21.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 1.4 | 38.4 | GO:0019835 | cytolysis(GO:0019835) |
| 1.1 | 3.3 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 1.0 | 8.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 1.0 | 12.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 1.0 | 5.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 1.0 | 14.2 | GO:0032094 | response to food(GO:0032094) |
| 1.0 | 2.0 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 1.0 | 9.5 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.9 | 61.4 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.9 | 2.7 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.9 | 5.3 | GO:1902023 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.8 | 19.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.8 | 13.4 | GO:0009409 | response to cold(GO:0009409) |
| 0.8 | 35.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.7 | 58.8 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.7 | 47.8 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.7 | 7.4 | GO:0045453 | bone resorption(GO:0045453) |
| 0.7 | 13.2 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.7 | 18.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.6 | 9.7 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.6 | 3.2 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.6 | 5.0 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.6 | 14.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.6 | 8.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.6 | 12.8 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.5 | 1.0 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.5 | 15.0 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.5 | 11.7 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.5 | 2.3 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.5 | 23.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.4 | 1.3 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.4 | 1.2 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.4 | 3.5 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.4 | 0.7 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.3 | 15.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.3 | 1.7 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.3 | 1.3 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.3 | 2.0 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.3 | 2.0 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.3 | 21.3 | GO:0006956 | complement activation(GO:0006956) protein activation cascade(GO:0072376) |
| 0.3 | 1.5 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.3 | 2.1 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.3 | 9.1 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.3 | 1.8 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 2.8 | GO:0021683 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 1.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.3 | 0.8 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.2 | 2.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.2 | 0.7 | GO:0006589 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.2 | 0.9 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 4.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.2 | 5.0 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.2 | 1.3 | GO:0007100 | mitotic centrosome separation(GO:0007100) centrosome separation(GO:0051299) |
| 0.2 | 3.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.2 | 8.6 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.2 | 1.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 | 3.0 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.2 | 3.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.6 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.1 | 2.8 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 2.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 2.0 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 9.5 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 11.1 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 0.4 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.1 | 0.6 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 3.9 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 0.6 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.9 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 6.7 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 2.0 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 10.4 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.1 | 3.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.5 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.1 | 5.1 | GO:0042310 | vasoconstriction(GO:0042310) |
| 0.1 | 4.3 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 0.9 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 6.2 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 0.8 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 0.9 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.1 | 0.8 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 4.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 2.3 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 0.4 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 1.0 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 7.7 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.1 | 9.1 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.1 | 3.7 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.1 | 1.1 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 1.9 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 5.8 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 0.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 1.2 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 2.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 2.5 | GO:0003309 | type B pancreatic cell differentiation(GO:0003309) |
| 0.1 | 4.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 1.8 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 7.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 1.9 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.1 | 12.2 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.1 | 0.5 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.1 | 2.6 | GO:0050727 | regulation of inflammatory response(GO:0050727) |
| 0.1 | 3.0 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.1 | 1.7 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.1 | 6.2 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.2 | GO:0061015 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.1 | 1.9 | GO:0007031 | peroxisome organization(GO:0007031) |
| 0.1 | 1.0 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 0.8 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 7.9 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.4 | GO:1902534 | macromitophagy(GO:0000423) lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) single-organism membrane invagination(GO:1902534) |
| 0.0 | 4.0 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.7 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 7.1 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 1.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 3.0 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.3 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.9 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.2 | GO:0035475 | angioblast cell migration involved in selective angioblast sprouting(GO:0035475) |
| 0.0 | 0.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.4 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 4.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 3.4 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 8.4 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.7 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.4 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.5 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.3 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.6 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.8 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 1.0 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.4 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.3 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.4 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 1.7 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 3.8 | GO:1904888 | cranial skeletal system development(GO:1904888) |
| 0.0 | 0.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 5.3 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 0.4 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.4 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 4.0 | 27.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 3.9 | 15.5 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 3.2 | 9.7 | GO:0043220 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 2.8 | 11.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 2.7 | 40.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 1.7 | 76.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 1.5 | 31.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 1.1 | 3.4 | GO:0031213 | RSF complex(GO:0031213) |
| 1.0 | 13.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.9 | 2.7 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.8 | 84.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.7 | 9.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.7 | 2.0 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.6 | 9.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.6 | 11.9 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.6 | 2.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.5 | 625.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.4 | 2.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.4 | 11.8 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.3 | 2.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 34.1 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.2 | 22.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.2 | 1.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 1.0 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.2 | 6.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 104.5 | GO:0005576 | extracellular region(GO:0005576) |
| 0.1 | 15.1 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.8 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 1.0 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 8.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.8 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.8 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 2.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 2.0 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 11.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 10.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 1.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 7.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.9 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 50.7 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.1 | 0.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 3.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 38.0 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 2.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 9.1 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 5.0 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 23.4 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 2.4 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.5 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 2.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 21.2 | 84.6 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 18.5 | 73.9 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 10.3 | 41.3 | GO:0070643 | vitamin D 25-hydroxylase activity(GO:0070643) |
| 10.2 | 61.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 7.2 | 28.9 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 5.4 | 16.3 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 4.3 | 12.8 | GO:0005499 | vitamin D binding(GO:0005499) |
| 4.0 | 465.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 3.8 | 11.5 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 3.4 | 24.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 3.1 | 125.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 2.9 | 11.7 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 2.8 | 11.2 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 2.5 | 10.1 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 2.1 | 6.4 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 2.0 | 9.8 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 1.7 | 5.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 1.6 | 13.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 1.6 | 11.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 1.6 | 12.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 1.5 | 21.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 1.3 | 6.7 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 1.3 | 35.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 1.3 | 5.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 1.1 | 10.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 1.0 | 19.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 1.0 | 9.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.8 | 2.5 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.8 | 37.5 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.8 | 3.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.8 | 5.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.7 | 16.9 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.7 | 2.7 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.6 | 9.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.6 | 1.9 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.6 | 3.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.6 | 11.3 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.6 | 3.1 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.6 | 1.8 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.6 | 141.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.6 | 13.6 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.6 | 7.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.5 | 2.1 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.5 | 13.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.5 | 5.9 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.4 | 6.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.4 | 3.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.4 | 3.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.4 | 1.2 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.4 | 2.3 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.3 | 1.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 4.7 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.3 | 10.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.3 | 14.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.3 | 11.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.3 | 0.8 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.3 | 1.6 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.3 | 0.8 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.2 | 14.1 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.2 | 0.7 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.2 | 0.7 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.2 | 0.9 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.2 | 6.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 0.9 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.2 | 3.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.2 | 20.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.2 | 0.8 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.2 | 2.0 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.2 | 17.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.2 | 4.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 21.7 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.2 | 4.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 8.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.2 | 3.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.2 | 4.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 2.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.6 | GO:0004945 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 4.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.8 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 7.3 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 4.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 6.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.0 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 2.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 7.6 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.8 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 6.1 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.1 | 3.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 1.1 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 2.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.4 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.1 | 41.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 3.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 0.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.9 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 20.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 3.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 7.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 3.5 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 7.6 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 11.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 11.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 8.2 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 18.9 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.4 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 1.0 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.1 | GO:0033745 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.0 | 3.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 1.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 2.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 2.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 66.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 3.3 | 147.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 1.6 | 241.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 1.5 | 18.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 1.3 | 10.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.6 | 10.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.5 | 13.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.4 | 19.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.3 | 7.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 1.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 1.7 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 2.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.3 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 0.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.4 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 0.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 2.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 2.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 2.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.3 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.2 | PID FOXO PATHWAY | FoxO family signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 92.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 6.8 | 68.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 4.4 | 66.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 4.4 | 30.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 4.2 | 54.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 3.7 | 84.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 3.5 | 62.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 2.1 | 17.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 2.0 | 17.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 1.9 | 54.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 1.6 | 24.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 1.6 | 72.9 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 1.2 | 13.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 1.1 | 28.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 1.1 | 9.5 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.9 | 9.4 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.8 | 38.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.7 | 4.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.6 | 10.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.5 | 6.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.5 | 35.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.4 | 2.9 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.3 | 4.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 1.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 2.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 2.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 1.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 1.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 24.9 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 8.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 7.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 2.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 2.0 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 1.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 3.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 0.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 0.9 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 2.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 3.2 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 0.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.9 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 2.1 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 1.0 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 1.3 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |