Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
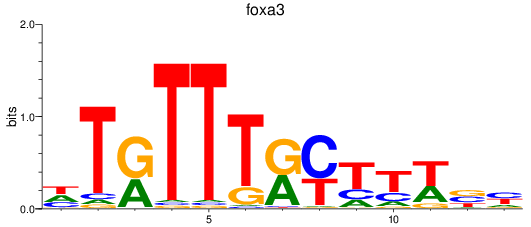
Results for foxa3
Z-value: 1.36
Transcription factors associated with foxa3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxa3
|
ENSDARG00000012788 | forkhead box A3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxa3 | dr11_v1_chr18_-_46354269_46354269 | 0.62 | 2.9e-11 | Click! |
Activity profile of foxa3 motif
Sorted Z-values of foxa3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_23978978 | 33.83 |
ENSDART00000058964
ENSDART00000135084 |
apoa2
|
apolipoprotein A-II |
| chr2_+_10134345 | 23.15 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr5_+_32345187 | 20.76 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr5_-_16351306 | 20.35 |
ENSDART00000168643
|
CABZ01088700.1
|
|
| chr5_+_28830643 | 19.55 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr5_+_28830388 | 18.81 |
ENSDART00000149150
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr20_+_6142433 | 18.45 |
ENSDART00000054084
ENSDART00000136986 |
ttr
|
transthyretin (prealbumin, amyloidosis type I) |
| chr5_+_28857969 | 17.02 |
ENSDART00000149850
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr11_+_13223625 | 17.02 |
ENSDART00000161275
|
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr21_-_1625976 | 16.39 |
ENSDART00000066621
|
vmo1b
|
vitelline membrane outer layer 1 homolog b |
| chr5_+_28848870 | 16.34 |
ENSDART00000149563
|
zgc:174259
|
zgc:174259 |
| chr2_-_38035235 | 16.31 |
ENSDART00000075904
|
cbln5
|
cerebellin 5 |
| chr5_+_28849155 | 16.28 |
ENSDART00000079090
|
zgc:174259
|
zgc:174259 |
| chr22_-_36856405 | 16.24 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr16_+_23913943 | 14.24 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr15_-_26552393 | 14.02 |
ENSDART00000150152
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr16_-_54455573 | 13.74 |
ENSDART00000075275
|
pklr
|
pyruvate kinase L/R |
| chr15_+_14856307 | 13.17 |
ENSDART00000167213
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr9_-_23253870 | 13.14 |
ENSDART00000143657
ENSDART00000169911 |
acmsd
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr5_+_28858345 | 12.45 |
ENSDART00000111180
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr15_-_26552652 | 12.22 |
ENSDART00000152336
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr4_-_6373735 | 12.05 |
ENSDART00000140100
|
MDFIC
|
si:ch73-156e19.1 |
| chr14_-_36799280 | 11.92 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr2_+_20539402 | 11.67 |
ENSDART00000129585
|
si:ch73-14h1.2
|
si:ch73-14h1.2 |
| chr18_+_35842933 | 11.41 |
ENSDART00000151587
ENSDART00000131121 |
ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr11_-_7380674 | 11.24 |
ENSDART00000014979
ENSDART00000103418 |
vtg3
|
vitellogenin 3, phosvitinless |
| chr6_-_13206255 | 11.04 |
ENSDART00000065373
|
eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr25_-_13188678 | 10.85 |
ENSDART00000125754
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr17_-_23709347 | 10.69 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr5_+_28797771 | 10.60 |
ENSDART00000188845
ENSDART00000149066 |
si:ch211-186e20.7
|
si:ch211-186e20.7 |
| chr18_-_16795262 | 10.29 |
ENSDART00000048722
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr2_+_37875789 | 9.81 |
ENSDART00000036318
ENSDART00000127679 |
cbln13
|
cerebellin 13 |
| chr20_+_15015557 | 9.66 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr23_+_26026383 | 9.65 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr12_-_26064480 | 9.44 |
ENSDART00000158215
ENSDART00000171206 ENSDART00000171212 ENSDART00000182956 ENSDART00000186779 |
ldb3b
|
LIM domain binding 3b |
| chr3_-_50865079 | 9.43 |
ENSDART00000164295
|
pmp22a
|
peripheral myelin protein 22a |
| chr13_+_22480857 | 8.82 |
ENSDART00000078721
ENSDART00000044719 ENSDART00000130957 ENSDART00000078757 ENSDART00000130424 ENSDART00000078747 |
ldb3a
|
LIM domain binding 3a |
| chr22_-_10110959 | 8.81 |
ENSDART00000031005
ENSDART00000147580 |
gls2b
|
glutaminase 2b (liver, mitochondrial) |
| chr8_+_49570884 | 8.75 |
ENSDART00000182117
ENSDART00000108613 |
rasef
|
RAS and EF-hand domain containing |
| chr14_+_32918484 | 8.74 |
ENSDART00000105721
|
lnx2b
|
ligand of numb-protein X 2b |
| chr16_-_17713859 | 8.32 |
ENSDART00000149275
|
zgc:174935
|
zgc:174935 |
| chr21_-_39081107 | 8.30 |
ENSDART00000075935
|
vtnb
|
vitronectin b |
| chr2_-_5475910 | 7.97 |
ENSDART00000100954
ENSDART00000172143 ENSDART00000132496 |
proca
proca
|
protein C (inactivator of coagulation factors Va and VIIIa), a protein C (inactivator of coagulation factors Va and VIIIa), a |
| chr16_-_21785261 | 7.86 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr21_-_25295087 | 7.82 |
ENSDART00000087910
ENSDART00000147860 |
st14b
|
suppression of tumorigenicity 14 (colon carcinoma) b |
| chr14_+_20893065 | 7.76 |
ENSDART00000079452
|
lygl1
|
lysozyme g-like 1 |
| chr6_+_52918537 | 7.59 |
ENSDART00000174229
|
or137-1
|
odorant receptor, family H, subfamily 137, member 1 |
| chr7_+_34305903 | 7.42 |
ENSDART00000173575
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr17_+_24036791 | 7.37 |
ENSDART00000140767
|
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr21_-_20840714 | 7.35 |
ENSDART00000144861
ENSDART00000139430 |
c6
|
complement component 6 |
| chr4_-_12795436 | 7.35 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr21_+_17768174 | 7.27 |
ENSDART00000141380
|
rxraa
|
retinoid X receptor, alpha a |
| chr23_+_27703749 | 7.08 |
ENSDART00000027224
|
lmbr1l
|
limb development membrane protein 1-like |
| chr1_+_36651059 | 6.95 |
ENSDART00000187475
|
ednraa
|
endothelin receptor type Aa |
| chr23_-_21515182 | 6.94 |
ENSDART00000142000
|
rnf207b
|
ring finger protein 207b |
| chr24_-_26369185 | 6.94 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr11_-_11336986 | 6.82 |
ENSDART00000016677
|
zgc:77929
|
zgc:77929 |
| chr18_+_17611627 | 6.73 |
ENSDART00000046891
|
cetp
|
cholesteryl ester transfer protein, plasma |
| chr8_+_39795918 | 6.54 |
ENSDART00000143413
|
si:ch211-170d8.2
|
si:ch211-170d8.2 |
| chr19_+_7636941 | 6.41 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr25_-_30344117 | 6.40 |
ENSDART00000167077
|
pdia3
|
protein disulfide isomerase family A, member 3 |
| chr21_-_35419486 | 6.40 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr3_+_12744083 | 6.29 |
ENSDART00000158554
ENSDART00000169545 |
cyp2k21
|
cytochrome P450, family 2, subfamily k, polypeptide 21 |
| chr17_+_30843881 | 6.07 |
ENSDART00000149600
ENSDART00000148547 |
tpp1
|
tripeptidyl peptidase I |
| chr14_-_4120636 | 6.06 |
ENSDART00000059230
|
irf2
|
interferon regulatory factor 2 |
| chr4_-_12795030 | 6.06 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr25_-_13188214 | 6.03 |
ENSDART00000187298
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr18_+_20566817 | 5.96 |
ENSDART00000100716
|
bida
|
BH3 interacting domain death agonist |
| chr5_-_66160415 | 5.94 |
ENSDART00000073895
|
mboat4
|
membrane bound O-acyltransferase domain containing 4 |
| chr7_+_13995792 | 5.81 |
ENSDART00000091470
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr24_-_31306724 | 5.78 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr6_-_8736766 | 5.77 |
ENSDART00000143956
|
cavin2b
|
caveolae associated protein 2b |
| chr3_+_12484008 | 5.74 |
ENSDART00000182229
|
vasnb
|
vasorin b |
| chr15_-_25365319 | 5.67 |
ENSDART00000152651
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr6_-_38930726 | 5.65 |
ENSDART00000154151
|
hdac7b
|
histone deacetylase 7b |
| chr20_+_52546186 | 5.61 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr3_+_23743139 | 5.56 |
ENSDART00000187409
|
hoxb3a
|
homeobox B3a |
| chr24_+_19542323 | 5.55 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr5_+_42092227 | 5.52 |
ENSDART00000097583
ENSDART00000171678 |
ubb
|
ubiquitin B |
| chr21_-_22951604 | 5.42 |
ENSDART00000083449
ENSDART00000180129 |
dub
|
duboraya |
| chr4_+_12358822 | 5.35 |
ENSDART00000172557
ENSDART00000150627 |
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr2_-_32262287 | 5.24 |
ENSDART00000056621
ENSDART00000039717 |
fam49ba
|
family with sequence similarity 49, member Ba |
| chr25_+_14087045 | 5.14 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr13_-_40726865 | 5.13 |
ENSDART00000099847
|
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr8_+_24747865 | 5.04 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr24_+_25258904 | 5.01 |
ENSDART00000155714
|
gabrr3b
|
gamma-aminobutyric acid (GABA) A receptor, rho 3b |
| chr6_-_53048291 | 4.99 |
ENSDART00000103267
|
fam212ab
|
family with sequence similarity 212, member Ab |
| chr10_-_15128771 | 4.86 |
ENSDART00000101261
|
spp1
|
secreted phosphoprotein 1 |
| chr19_-_46091497 | 4.83 |
ENSDART00000178772
ENSDART00000167255 |
ptdss1b
si:dkey-108k24.2
|
phosphatidylserine synthase 1b si:dkey-108k24.2 |
| chr2_+_25839940 | 4.81 |
ENSDART00000139927
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr7_-_5029478 | 4.78 |
ENSDART00000193819
|
ltb4r
|
leukotriene B4 receptor |
| chr2_+_25840463 | 4.72 |
ENSDART00000125178
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr4_-_1914228 | 4.69 |
ENSDART00000087835
|
ano6
|
anoctamin 6 |
| chr1_-_44940830 | 4.64 |
ENSDART00000097500
ENSDART00000134464 ENSDART00000137216 |
tmem176
|
transmembrane protein 176 |
| chr3_+_29941777 | 4.61 |
ENSDART00000113889
|
ifi35
|
interferon-induced protein 35 |
| chr20_-_9980318 | 4.60 |
ENSDART00000080664
|
ACTC1
|
zgc:86709 |
| chr2_+_23731194 | 4.57 |
ENSDART00000155747
|
slc22a13a
|
solute carrier family 22 member 13a |
| chr3_-_41292569 | 4.53 |
ENSDART00000111856
|
sdk1a
|
sidekick cell adhesion molecule 1a |
| chr16_+_11724230 | 4.49 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr9_-_29497916 | 4.47 |
ENSDART00000060246
|
dnajc3a
|
DnaJ (Hsp40) homolog, subfamily C, member 3a |
| chr17_+_8799661 | 4.42 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr6_-_25384526 | 4.38 |
ENSDART00000160544
|
PKN2 (1 of many)
|
zgc:153916 |
| chr3_-_6719232 | 4.25 |
ENSDART00000154294
|
atg4db
|
autophagy related 4D, cysteine peptidase b |
| chr20_+_51104367 | 4.23 |
ENSDART00000073981
|
eif2s1b
|
eukaryotic translation initiation factor 2, subunit 1 alpha b |
| chr22_+_7497319 | 4.15 |
ENSDART00000034564
|
CELA1 (1 of many)
|
zgc:92511 |
| chr4_+_14981854 | 4.15 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr18_-_44847855 | 4.11 |
ENSDART00000086823
|
srpr
|
signal recognition particle receptor (docking protein) |
| chr3_+_32571929 | 4.09 |
ENSDART00000151025
|
si:ch73-248e21.1
|
si:ch73-248e21.1 |
| chr24_-_26485098 | 4.06 |
ENSDART00000135496
ENSDART00000009609 ENSDART00000133782 ENSDART00000141029 ENSDART00000113739 |
eif5a
|
eukaryotic translation initiation factor 5A |
| chr16_+_36748538 | 3.93 |
ENSDART00000139069
|
decr1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr23_+_42813415 | 3.89 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr1_+_36612660 | 3.87 |
ENSDART00000190784
|
ednraa
|
endothelin receptor type Aa |
| chr23_-_44848961 | 3.86 |
ENSDART00000136839
|
wu:fb72h05
|
wu:fb72h05 |
| chr14_-_6931889 | 3.86 |
ENSDART00000166439
|
si:ch211-266k2.1
|
si:ch211-266k2.1 |
| chr17_-_42988356 | 3.85 |
ENSDART00000024558
|
zgc:92137
|
zgc:92137 |
| chr8_-_43923788 | 3.75 |
ENSDART00000146152
|
adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr21_+_37436907 | 3.75 |
ENSDART00000182611
ENSDART00000076328 |
pgrmc1
|
progesterone receptor membrane component 1 |
| chr7_+_58730201 | 3.72 |
ENSDART00000073640
|
plag1
|
pleiomorphic adenoma gene 1 |
| chr2_+_38161318 | 3.70 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr5_-_54712159 | 3.68 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr7_+_4682659 | 3.65 |
ENSDART00000112697
|
si:ch211-225k7.2
|
si:ch211-225k7.2 |
| chr7_+_20512419 | 3.60 |
ENSDART00000173907
|
si:dkey-19b23.14
|
si:dkey-19b23.14 |
| chr10_+_23553533 | 3.57 |
ENSDART00000159654
|
CR847844.2
|
|
| chr15_-_44052927 | 3.53 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr4_+_17310143 | 3.53 |
ENSDART00000004717
|
igf1
|
insulin-like growth factor 1 |
| chr1_+_9153141 | 3.50 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr11_-_30634286 | 3.48 |
ENSDART00000191019
|
zgc:153665
|
zgc:153665 |
| chr9_-_34396264 | 3.47 |
ENSDART00000045754
|
grtp1b
|
growth hormone regulated TBC protein 1b |
| chr10_-_11385155 | 3.46 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr10_+_15603082 | 3.42 |
ENSDART00000024450
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr7_+_13830052 | 3.39 |
ENSDART00000191360
|
abhd2a
|
abhydrolase domain containing 2a |
| chr3_+_15828999 | 3.39 |
ENSDART00000104397
|
tmem11
|
transmembrane protein 11 |
| chr15_+_15390882 | 3.39 |
ENSDART00000062024
|
ca4b
|
carbonic anhydrase IV b |
| chr15_-_25571865 | 3.37 |
ENSDART00000077836
|
mmp20b
|
matrix metallopeptidase 20b (enamelysin) |
| chr21_+_30549512 | 3.36 |
ENSDART00000132831
|
rab38c
|
RAB38c, member of RAS oncogene family |
| chr7_-_51300277 | 3.34 |
ENSDART00000174174
|
gc2
|
guanylyl cyclase 2 |
| chr11_+_21137948 | 3.33 |
ENSDART00000110673
|
il10
|
interleukin 10 |
| chr13_+_46803979 | 3.32 |
ENSDART00000159260
|
CU695232.1
|
|
| chr5_-_32292965 | 3.31 |
ENSDART00000183522
ENSDART00000131983 |
myhz1.2
|
myosin, heavy polypeptide 1.2, skeletal muscle |
| chr1_+_1805294 | 3.28 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 3 |
| chr3_+_2719243 | 3.27 |
ENSDART00000189256
|
CR388047.3
|
|
| chr7_+_48705227 | 3.23 |
ENSDART00000174034
|
AL929208.1
|
|
| chr24_-_32025637 | 3.19 |
ENSDART00000180448
ENSDART00000159034 |
rsu1
|
Ras suppressor protein 1 |
| chr21_-_17296789 | 3.18 |
ENSDART00000192180
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr11_+_2391469 | 3.13 |
ENSDART00000182121
|
igfbp6a
|
insulin-like growth factor binding protein 6a |
| chr3_-_43770876 | 3.13 |
ENSDART00000160162
|
zgc:92162
|
zgc:92162 |
| chr5_-_20194876 | 3.11 |
ENSDART00000122587
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr12_-_2993095 | 3.10 |
ENSDART00000152316
|
si:dkey-202c14.3
|
si:dkey-202c14.3 |
| chr3_+_32129632 | 3.07 |
ENSDART00000174522
|
zgc:109934
|
zgc:109934 |
| chr15_-_37719679 | 3.07 |
ENSDART00000184025
|
si:dkey-117a8.1
|
si:dkey-117a8.1 |
| chr12_-_29301022 | 3.04 |
ENSDART00000187826
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr13_-_47403154 | 3.04 |
ENSDART00000114318
|
bcl2l11
|
BCL2 like 11 |
| chr7_-_32020100 | 3.03 |
ENSDART00000185433
|
kif18a
|
kinesin family member 18A |
| chr22_-_14128716 | 3.02 |
ENSDART00000140323
|
si:ch211-246m6.4
|
si:ch211-246m6.4 |
| chr11_-_183328 | 3.02 |
ENSDART00000168562
|
avil
|
advillin |
| chr8_-_3312384 | 3.02 |
ENSDART00000035965
|
fut9b
|
fucosyltransferase 9b |
| chr8_+_30456161 | 3.01 |
ENSDART00000085894
|
pgm5
|
phosphoglucomutase 5 |
| chr16_-_35952789 | 3.00 |
ENSDART00000180118
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr4_-_30055196 | 2.99 |
ENSDART00000139539
|
si:rp71-7l19.2
|
si:rp71-7l19.2 |
| chr7_-_17570923 | 2.99 |
ENSDART00000188476
ENSDART00000080624 |
nitr5
|
novel immune-type receptor 5 |
| chr5_-_34875858 | 2.99 |
ENSDART00000085086
|
arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr15_+_36187434 | 2.97 |
ENSDART00000181536
ENSDART00000099501 ENSDART00000154432 |
masp1
|
mannan-binding lectin serine peptidase 1 |
| chr15_-_8191841 | 2.94 |
ENSDART00000156663
|
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr17_-_53439866 | 2.94 |
ENSDART00000154826
|
mycbp
|
c-myc binding protein |
| chr16_-_25233515 | 2.93 |
ENSDART00000058943
|
zgc:110182
|
zgc:110182 |
| chr3_-_55525627 | 2.91 |
ENSDART00000189234
|
tex2
|
testis expressed 2 |
| chr19_-_19720744 | 2.90 |
ENSDART00000170636
|
evx1
|
even-skipped homeobox 1 |
| chr7_-_13906409 | 2.87 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr3_+_13559199 | 2.85 |
ENSDART00000166547
|
si:ch73-106n3.1
|
si:ch73-106n3.1 |
| chr17_+_4400738 | 2.84 |
ENSDART00000170204
|
si:zfos-364h11.1
|
si:zfos-364h11.1 |
| chr23_-_21215311 | 2.83 |
ENSDART00000112424
|
megf6a
|
multiple EGF-like-domains 6a |
| chr2_-_27330181 | 2.81 |
ENSDART00000137053
|
tmx3a
|
thioredoxin related transmembrane protein 3a |
| chr16_-_13992646 | 2.81 |
ENSDART00000139623
|
si:dkey-85k15.6
|
si:dkey-85k15.6 |
| chr11_+_2391649 | 2.80 |
ENSDART00000104571
|
igfbp6a
|
insulin-like growth factor binding protein 6a |
| chr17_+_33418475 | 2.78 |
ENSDART00000169145
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr15_-_669476 | 2.77 |
ENSDART00000153687
ENSDART00000030603 |
si:ch211-210b2.2
|
si:ch211-210b2.2 |
| chr11_-_25539323 | 2.77 |
ENSDART00000155785
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr8_-_25690966 | 2.74 |
ENSDART00000033701
|
sema3ga
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ga |
| chr22_+_24673168 | 2.73 |
ENSDART00000135257
|
si:rp71-23d18.8
|
si:rp71-23d18.8 |
| chr8_-_31701157 | 2.72 |
ENSDART00000141799
|
fbxo4
|
F-box protein 4 |
| chr3_-_41292275 | 2.70 |
ENSDART00000144088
|
sdk1a
|
sidekick cell adhesion molecule 1a |
| chr3_-_56871330 | 2.68 |
ENSDART00000014103
|
zgc:112148
|
zgc:112148 |
| chr19_+_30867845 | 2.67 |
ENSDART00000047461
|
mfsd2ab
|
major facilitator superfamily domain containing 2ab |
| chr15_+_24644016 | 2.66 |
ENSDART00000043292
|
smtnl
|
smoothelin, like |
| chr3_+_32492467 | 2.66 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr16_-_38001040 | 2.63 |
ENSDART00000133861
ENSDART00000138711 ENSDART00000143846 ENSDART00000146564 |
si:ch211-198c19.3
|
si:ch211-198c19.3 |
| chr8_+_30780985 | 2.62 |
ENSDART00000193971
ENSDART00000111533 |
tas2r200.2
|
taste receptor, type 2, member 200, tandem duplicate 2 |
| chr5_+_29726428 | 2.60 |
ENSDART00000143183
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr11_+_1584747 | 2.57 |
ENSDART00000154583
|
si:dkey-40c23.2
|
si:dkey-40c23.2 |
| chr7_-_58729894 | 2.55 |
ENSDART00000149347
|
chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr1_+_49668423 | 2.54 |
ENSDART00000150880
|
tsga10
|
testis specific, 10 |
| chr25_+_7982979 | 2.53 |
ENSDART00000171904
|
ucmab
|
upper zone of growth plate and cartilage matrix associated b |
| chr23_-_29357764 | 2.53 |
ENSDART00000156512
|
si:ch211-129o18.4
|
si:ch211-129o18.4 |
| chr13_+_40692804 | 2.51 |
ENSDART00000109822
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr3_+_5575313 | 2.50 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr8_-_20245892 | 2.50 |
ENSDART00000136911
|
acer1
|
alkaline ceramidase 1 |
| chr5_-_14509137 | 2.48 |
ENSDART00000180742
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr8_-_19313510 | 2.45 |
ENSDART00000164780
ENSDART00000137133 |
rgl1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr8_-_27858458 | 2.43 |
ENSDART00000132632
ENSDART00000136562 |
cttnbp2nlb
|
CTTNBP2 N-terminal like b |
| chr6_+_13206516 | 2.41 |
ENSDART00000036927
|
ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxa3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.9 | 20.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 4.3 | 17.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 3.6 | 10.7 | GO:0050427 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 2.6 | 13.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 2.6 | 7.8 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 2.3 | 16.2 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 2.3 | 6.9 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 2.2 | 6.7 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 2.2 | 13.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) quinolinate metabolic process(GO:0046874) |
| 1.9 | 13.6 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 1.9 | 157.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 1.8 | 5.4 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 1.8 | 10.8 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 1.7 | 16.7 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 1.6 | 4.7 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 1.5 | 6.0 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 1.2 | 3.7 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 1.2 | 13.4 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 1.1 | 8.8 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 1.1 | 7.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.9 | 2.8 | GO:0010359 | regulation of anion channel activity(GO:0010359) |
| 0.9 | 2.7 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.9 | 3.4 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.8 | 3.1 | GO:0009080 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.7 | 2.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.7 | 4.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.7 | 10.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.7 | 2.2 | GO:0045141 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.7 | 2.2 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.7 | 2.8 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.7 | 5.5 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.7 | 3.4 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.7 | 3.4 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.6 | 1.9 | GO:0045601 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of endothelial cell differentiation(GO:0045601) |
| 0.6 | 2.5 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.6 | 1.8 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.6 | 7.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.6 | 9.7 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.5 | 2.2 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.5 | 5.9 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.5 | 2.7 | GO:0045056 | transcytosis(GO:0045056) |
| 0.5 | 7.4 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.5 | 5.8 | GO:0045453 | bone resorption(GO:0045453) |
| 0.5 | 7.1 | GO:0032094 | response to food(GO:0032094) |
| 0.5 | 1.5 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.5 | 2.0 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.5 | 2.0 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.5 | 1.9 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.5 | 4.8 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.4 | 35.8 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.4 | 6.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.4 | 5.8 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.4 | 3.7 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.4 | 3.7 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.4 | 1.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.4 | 3.1 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.4 | 1.9 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.4 | 1.8 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.3 | 2.0 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.3 | 8.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.3 | 11.2 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.3 | 2.9 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) embryonic skeletal joint development(GO:0072498) |
| 0.3 | 16.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.3 | 1.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.3 | 7.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.3 | 1.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.3 | 1.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.2 | 2.0 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 2.6 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.2 | 2.5 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.2 | 0.9 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 | 2.9 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.2 | 5.7 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.2 | 1.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.2 | 13.7 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.2 | 5.6 | GO:1903039 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.2 | 4.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.2 | 0.6 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.2 | 2.5 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.2 | 1.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 3.4 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.2 | 0.9 | GO:0050996 | positive regulation of lipid catabolic process(GO:0050996) |
| 0.2 | 5.4 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.2 | 3.3 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 2.5 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.2 | 1.8 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.2 | 1.3 | GO:0030320 | cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 | 1.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.2 | 1.3 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.2 | 4.3 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.2 | 3.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 2.9 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 1.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.6 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 3.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.9 | GO:0009217 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.1 | 2.1 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 | 0.4 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 1.5 | GO:0048938 | myelination of lateral line nerve axons(GO:0048897) posterior lateral line nerve glial cell differentiation(GO:0048931) myelination of posterior lateral line nerve axons(GO:0048932) lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048938) posterior lateral line nerve glial cell development(GO:0048941) posterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048942) |
| 0.1 | 1.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 8.0 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 7.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.6 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.1 | 1.2 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 2.8 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 9.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 0.9 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 2.0 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 7.1 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 0.5 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 13.8 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.1 | 0.4 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.1 | 7.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 1.6 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 5.0 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 2.7 | GO:2001236 | regulation of extrinsic apoptotic signaling pathway(GO:2001236) |
| 0.1 | 0.9 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.6 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 2.3 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 3.3 | GO:0032496 | response to lipopolysaccharide(GO:0032496) |
| 0.1 | 2.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 1.8 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 | 4.9 | GO:0060348 | bone development(GO:0060348) |
| 0.1 | 0.6 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 9.2 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.1 | 3.5 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 1.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 0.7 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 3.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 2.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 3.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.5 | GO:0046958 | learning(GO:0007612) nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 3.9 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 7.1 | GO:0034976 | response to endoplasmic reticulum stress(GO:0034976) |
| 0.1 | 2.4 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 9.9 | GO:0014032 | neural crest cell development(GO:0014032) |
| 0.0 | 1.7 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 3.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 3.0 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.6 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 8.1 | GO:0045597 | positive regulation of cell differentiation(GO:0045597) |
| 0.0 | 2.2 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 1.2 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 2.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 7.0 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.7 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.0 | 3.0 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.0 | 1.0 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 3.5 | GO:0009953 | dorsal/ventral pattern formation(GO:0009953) |
| 0.0 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 1.6 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 7.2 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.6 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 2.5 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.9 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 7.9 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 1.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.3 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.7 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 8.7 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 7.2 | GO:0019941 | modification-dependent protein catabolic process(GO:0019941) |
| 0.0 | 0.9 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 2.6 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.9 | 20.8 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 2.4 | 9.6 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 2.4 | 16.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 2.2 | 13.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 1.4 | 4.2 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 1.2 | 3.7 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.8 | 3.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.8 | 3.0 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.6 | 4.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.6 | 2.5 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.6 | 8.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.6 | 1.7 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.5 | 2.0 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.5 | 7.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 1.2 | GO:0043202 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.4 | 18.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.4 | 1.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.3 | 1.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 9.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 7.1 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.2 | 11.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.2 | 1.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 11.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.2 | 203.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.2 | 0.9 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 4.2 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 5.8 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 5.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 6.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 12.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.0 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 3.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 4.3 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 1.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 10.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 1.0 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 3.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 6.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 1.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.5 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 20.6 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 1.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 7.1 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 2.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.6 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.7 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 1.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 4.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 2.2 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 8.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 4.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 20.2 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.1 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 0.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 10.7 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 1.1 | GO:0043025 | neuronal cell body(GO:0043025) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 18.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 4.3 | 17.0 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 3.6 | 10.7 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 2.5 | 7.4 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 2.3 | 13.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.9 | 7.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 1.7 | 5.1 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 1.6 | 11.2 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 1.5 | 10.8 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 1.2 | 137.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 1.0 | 4.2 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 1.0 | 39.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 1.0 | 8.8 | GO:0004359 | glutaminase activity(GO:0004359) |
| 1.0 | 5.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.9 | 10.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.9 | 18.4 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.8 | 3.1 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.7 | 5.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.7 | 2.2 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.7 | 2.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.7 | 4.8 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.7 | 30.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.7 | 5.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.6 | 5.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.6 | 1.7 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.6 | 3.4 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.6 | 2.8 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.5 | 5.9 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.5 | 7.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.5 | 6.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.5 | 6.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.5 | 6.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.4 | 2.5 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.4 | 2.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.4 | 1.2 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.3 | 5.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.3 | 3.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 5.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 2.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 5.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.3 | 0.9 | GO:0070186 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.3 | 1.6 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.3 | 8.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 13.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.3 | 3.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 2.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.3 | 6.7 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.3 | 1.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.3 | 3.0 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.2 | 7.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 1.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 2.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 1.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 1.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 1.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.2 | 0.5 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.9 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 3.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 0.6 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 4.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 28.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 3.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 4.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 2.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 2.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.7 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 2.0 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 6.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 3.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 5.9 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.1 | 4.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.9 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.1 | 2.0 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 3.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 2.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 4.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 1.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 2.7 | GO:0099604 | ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 2.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 2.7 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.1 | 2.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.2 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 3.6 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 3.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 2.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.1 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.6 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 1.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 22.7 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 1.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 2.6 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 5.4 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.6 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 18.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 5.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 16.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.7 | 32.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.4 | 13.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.4 | 3.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.4 | 4.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 37.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 7.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 1.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.2 | 3.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.2 | 1.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 2.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 12.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 4.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 6.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 2.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 3.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 2.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.8 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 1.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.8 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.1 | 2.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 1.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 34.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 2.7 | 13.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 1.2 | 31.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 1.1 | 23.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 1.1 | 16.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.0 | 13.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.9 | 18.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.7 | 6.7 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.6 | 6.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.5 | 13.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.5 | 4.8 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.4 | 7.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.4 | 6.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.4 | 10.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.4 | 2.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.3 | 4.9 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.3 | 3.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 3.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.3 | 10.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 3.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 1.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.2 | 2.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 2.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.3 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 1.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 2.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 11.0 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 2.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.8 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 2.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 2.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |