Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
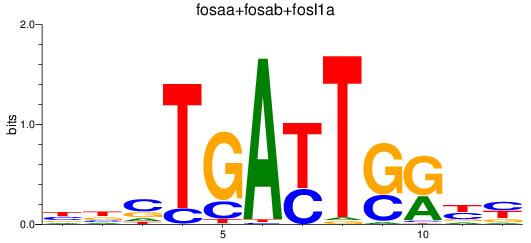
Results for fosaa+fosab+fosl1a
Z-value: 2.10
Transcription factors associated with fosaa+fosab+fosl1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
fosl1a
|
ENSDARG00000015355 | FOS-like antigen 1a |
|
fosab
|
ENSDARG00000031683 | v-fos FBJ murine osteosarcoma viral oncogene homolog Ab |
|
fosaa
|
ENSDARG00000040135 | v-fos FBJ murine osteosarcoma viral oncogene homolog Aa |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| fosaa | dr11_v1_chr17_-_50234004_50234004 | -0.15 | 1.5e-01 | Click! |
| fosl1a | dr11_v1_chr14_-_30747686_30747686 | 0.12 | 2.6e-01 | Click! |
| fosab | dr11_v1_chr20_-_46554440_46554440 | 0.00 | 9.9e-01 | Click! |
Activity profile of fosaa+fosab+fosl1a motif
Sorted Z-values of fosaa+fosab+fosl1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_44883805 | 62.95 |
ENSDART00000182805
|
si:ch73-361h17.1
|
si:ch73-361h17.1 |
| chr17_-_6514962 | 17.41 |
ENSDART00000163514
|
dnajc5gb
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma b |
| chr13_+_2394534 | 14.83 |
ENSDART00000172535
ENSDART00000006990 |
elovl5
|
ELOVL fatty acid elongase 5 |
| chr1_-_56080112 | 14.57 |
ENSDART00000075469
ENSDART00000161473 |
c3a.6
|
complement component c3a, duplicate 6 |
| chr1_-_20928772 | 14.09 |
ENSDART00000078277
|
msmo1
|
methylsterol monooxygenase 1 |
| chr19_+_14109348 | 13.62 |
ENSDART00000159015
|
zgc:175136
|
zgc:175136 |
| chr7_+_34794829 | 13.20 |
ENSDART00000009698
ENSDART00000075089 ENSDART00000173456 |
esrp2
|
epithelial splicing regulatory protein 2 |
| chr22_+_38173960 | 12.93 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr11_+_29537756 | 12.89 |
ENSDART00000103388
|
wu:fi42e03
|
wu:fi42e03 |
| chr24_+_17334682 | 12.24 |
ENSDART00000018868
|
pdia4
|
protein disulfide isomerase family A, member 4 |
| chr24_+_10027902 | 11.67 |
ENSDART00000175961
ENSDART00000172773 |
si:ch211-146l10.8
|
si:ch211-146l10.8 |
| chr1_-_33647138 | 11.38 |
ENSDART00000142111
ENSDART00000015547 |
cldng
|
claudin g |
| chr22_+_32228882 | 11.06 |
ENSDART00000092082
|
manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr10_+_6010570 | 10.95 |
ENSDART00000190025
ENSDART00000163680 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr18_-_5595546 | 10.24 |
ENSDART00000191825
|
cyp1a
|
cytochrome P450, family 1, subfamily A |
| chr6_+_60055168 | 9.74 |
ENSDART00000008752
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr3_-_25377163 | 9.55 |
ENSDART00000055490
|
kpna2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr5_-_2721686 | 9.46 |
ENSDART00000169404
|
hspa5
|
heat shock protein 5 |
| chr20_-_7176809 | 9.26 |
ENSDART00000012247
ENSDART00000125019 |
dhcr24
|
24-dehydrocholesterol reductase |
| chr13_+_2394264 | 9.18 |
ENSDART00000168595
|
elovl5
|
ELOVL fatty acid elongase 5 |
| chr21_+_3897680 | 8.82 |
ENSDART00000170653
|
dolpp1
|
dolichyldiphosphatase 1 |
| chr18_+_13164325 | 8.73 |
ENSDART00000189057
|
tat
|
tyrosine aminotransferase |
| chr25_-_4148719 | 8.66 |
ENSDART00000112880
ENSDART00000023278 |
fads2
|
fatty acid desaturase 2 |
| chr1_-_54706039 | 8.58 |
ENSDART00000083633
|
exosc1
|
exosome component 1 |
| chr24_-_10006158 | 8.54 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr3_-_32965848 | 8.53 |
ENSDART00000050930
|
casp6
|
caspase 6, apoptosis-related cysteine peptidase |
| chr14_-_14659023 | 8.45 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr2_-_55797318 | 8.45 |
ENSDART00000158147
|
calr3b
|
calreticulin 3b |
| chr20_-_52939501 | 8.33 |
ENSDART00000166508
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr3_-_40276057 | 8.20 |
ENSDART00000132225
ENSDART00000074737 |
shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr22_-_22719440 | 7.94 |
ENSDART00000166794
|
nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr19_+_791538 | 7.74 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr7_+_30626378 | 7.45 |
ENSDART00000173533
ENSDART00000052541 |
ccnb2
|
cyclin B2 |
| chr12_+_48390715 | 7.44 |
ENSDART00000149351
|
scd
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr6_-_609880 | 7.36 |
ENSDART00000149248
ENSDART00000148867 ENSDART00000149414 ENSDART00000148552 ENSDART00000148391 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr18_+_62932 | 7.34 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr20_-_30931139 | 7.27 |
ENSDART00000006778
ENSDART00000146376 |
acat2
|
acetyl-CoA acetyltransferase 2 |
| chr23_-_41651759 | 7.27 |
ENSDART00000146808
|
si:ch73-184c24.1
|
si:ch73-184c24.1 |
| chr18_-_5598958 | 7.18 |
ENSDART00000161538
|
cyp1a
|
cytochrome P450, family 1, subfamily A |
| chr25_-_13728111 | 7.10 |
ENSDART00000169865
|
lcat
|
lecithin-cholesterol acyltransferase |
| chr5_-_54712159 | 7.03 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr17_+_19481049 | 6.95 |
ENSDART00000024194
|
kif11
|
kinesin family member 11 |
| chr24_-_2843107 | 6.79 |
ENSDART00000165290
|
cyb5a
|
cytochrome b5 type A (microsomal) |
| chr24_-_9997948 | 6.77 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr3_-_4663602 | 6.73 |
ENSDART00000083532
|
slc25a38a
|
solute carrier family 25, member 38a |
| chr24_-_9989634 | 6.72 |
ENSDART00000115275
|
zgc:152652
|
zgc:152652 |
| chr2_-_32643738 | 6.67 |
ENSDART00000112452
|
si:dkeyp-73d8.9
|
si:dkeyp-73d8.9 |
| chr12_+_6041575 | 6.66 |
ENSDART00000091868
|
g6pca.2
|
glucose-6-phosphatase a, catalytic subunit, tandem duplicate 2 |
| chr16_+_26012569 | 6.59 |
ENSDART00000148846
|
prss59.1
|
protease, serine, 59, tandem duplicate 1 |
| chr15_+_478524 | 6.54 |
ENSDART00000018062
|
si:ch1073-280e3.1
|
si:ch1073-280e3.1 |
| chr2_-_47620806 | 6.53 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr25_-_6261693 | 6.52 |
ENSDART00000135808
|
ireb2
|
iron-responsive element binding protein 2 |
| chr21_-_5077715 | 6.50 |
ENSDART00000081954
|
haus1
|
HAUS augmin-like complex, subunit 1 |
| chr20_-_2641233 | 6.47 |
ENSDART00000145335
ENSDART00000133121 |
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr10_+_26972755 | 6.42 |
ENSDART00000042162
|
tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr13_-_31397987 | 6.37 |
ENSDART00000008287
|
pgam1a
|
phosphoglycerate mutase 1a |
| chr2_-_43653015 | 6.26 |
ENSDART00000148454
|
itgb1b.2
|
integrin, beta 1b.2 |
| chr5_-_50992690 | 6.23 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr10_+_17026870 | 6.17 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr20_-_15089738 | 6.13 |
ENSDART00000164552
|
si:dkey-239i20.2
|
si:dkey-239i20.2 |
| chr12_-_48671612 | 6.12 |
ENSDART00000007202
|
zgc:92749
|
zgc:92749 |
| chr17_+_12159529 | 6.11 |
ENSDART00000046802
|
sccpdha
|
saccharopine dehydrogenase a |
| chr19_-_617246 | 6.11 |
ENSDART00000062551
|
cyp51
|
cytochrome P450, family 51 |
| chr17_+_20923691 | 6.08 |
ENSDART00000122407
|
cdk1
|
cyclin-dependent kinase 1 |
| chr6_-_55297274 | 6.07 |
ENSDART00000184283
|
ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr1_-_18811517 | 6.04 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr23_-_44723102 | 6.03 |
ENSDART00000129138
|
mogat3a
|
monoacylglycerol O-acyltransferase 3a |
| chr16_+_46695777 | 5.95 |
ENSDART00000169767
|
rab25b
|
RAB25, member RAS oncogene family b |
| chr4_+_1530287 | 5.94 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
| chr8_+_19514294 | 5.92 |
ENSDART00000170622
|
si:ch73-281k2.5
|
si:ch73-281k2.5 |
| chr25_+_5983430 | 5.89 |
ENSDART00000074814
|
ppib
|
peptidylprolyl isomerase B (cyclophilin B) |
| chr17_-_15149192 | 5.83 |
ENSDART00000180511
ENSDART00000103405 |
gch1
|
GTP cyclohydrolase 1 |
| chr24_-_9979342 | 5.81 |
ENSDART00000138576
ENSDART00000191206 |
zgc:171977
|
zgc:171977 |
| chr10_+_26973063 | 5.78 |
ENSDART00000143162
ENSDART00000186210 |
tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr8_+_14886452 | 5.77 |
ENSDART00000146589
|
soat1
|
sterol O-acyltransferase 1 |
| chr8_-_2616326 | 5.71 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr22_-_10539180 | 5.64 |
ENSDART00000131217
|
ippk
|
inositol 1,3,4,5,6-pentakisphosphate 2-kinase |
| chr3_-_49514874 | 5.63 |
ENSDART00000167179
|
asf1ba
|
anti-silencing function 1Ba histone chaperone |
| chr10_-_25699454 | 5.63 |
ENSDART00000064376
|
sod1
|
superoxide dismutase 1, soluble |
| chr18_+_3572314 | 5.61 |
ENSDART00000169814
ENSDART00000157819 |
serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr3_+_1167026 | 5.52 |
ENSDART00000031823
ENSDART00000155340 |
triobpb
|
TRIO and F-actin binding protein b |
| chr13_-_25198025 | 5.50 |
ENSDART00000159585
ENSDART00000144227 |
adka
|
adenosine kinase a |
| chr9_+_38481780 | 5.46 |
ENSDART00000087241
|
lss
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr9_-_29497916 | 5.44 |
ENSDART00000060246
|
dnajc3a
|
DnaJ (Hsp40) homolog, subfamily C, member 3a |
| chr12_-_42368296 | 5.42 |
ENSDART00000171075
|
zgc:111868
|
zgc:111868 |
| chr21_+_26733529 | 5.41 |
ENSDART00000168379
|
pcxa
|
pyruvate carboxylase a |
| chr5_+_28857969 | 5.36 |
ENSDART00000149850
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr1_+_29068654 | 5.32 |
ENSDART00000053932
|
cbsa
|
cystathionine-beta-synthase a |
| chr2_-_43653328 | 5.32 |
ENSDART00000037808
|
itgb1b.2
|
integrin, beta 1b.2 |
| chr25_+_10416583 | 5.30 |
ENSDART00000073907
|
ehf
|
ets homologous factor |
| chr3_-_32541033 | 5.29 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr6_+_3710865 | 5.21 |
ENSDART00000170781
|
phospho2
|
phosphatase, orphan 2 |
| chr10_+_29770120 | 5.18 |
ENSDART00000100032
ENSDART00000193205 |
hyou1
|
hypoxia up-regulated 1 |
| chr5_+_1493767 | 5.14 |
ENSDART00000022132
|
haus4
|
HAUS augmin-like complex, subunit 4 |
| chr22_-_17611742 | 5.12 |
ENSDART00000144031
|
gpx4a
|
glutathione peroxidase 4a |
| chr8_-_16609004 | 5.12 |
ENSDART00000102556
|
tor3a
|
torsin family 3, member A |
| chr20_+_48116476 | 5.06 |
ENSDART00000043938
|
tram2
|
translocation associated membrane protein 2 |
| chr24_-_9991153 | 5.00 |
ENSDART00000137794
ENSDART00000106252 ENSDART00000188309 ENSDART00000188266 ENSDART00000188660 ENSDART00000185713 ENSDART00000179773 |
zgc:152652
|
zgc:152652 |
| chr12_+_46543572 | 5.00 |
ENSDART00000167510
|
hid1b
|
HID1 domain containing b |
| chr13_-_8692860 | 4.99 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr7_-_41014773 | 4.96 |
ENSDART00000013785
|
insig1
|
insulin induced gene 1 |
| chr16_-_12060770 | 4.95 |
ENSDART00000183237
ENSDART00000103948 |
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr14_-_16476863 | 4.85 |
ENSDART00000089021
|
canx
|
calnexin |
| chr5_-_33022014 | 4.85 |
ENSDART00000061149
|
zgc:55461
|
zgc:55461 |
| chr19_+_33464688 | 4.84 |
ENSDART00000142275
|
triqk
|
triple QxxK/R motif containing |
| chr12_+_23424108 | 4.83 |
ENSDART00000077732
|
bambia
|
BMP and activin membrane-bound inhibitor (Xenopus laevis) homolog a |
| chr2_+_15612755 | 4.82 |
ENSDART00000003035
|
amy2a
|
amylase, alpha 2A (pancreatic) |
| chr24_+_12835935 | 4.73 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr21_+_6114709 | 4.73 |
ENSDART00000065858
|
fpgs
|
folylpolyglutamate synthase |
| chr8_-_38201415 | 4.72 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr23_-_46040618 | 4.71 |
ENSDART00000161415
|
CABZ01080918.1
|
|
| chr10_+_3520256 | 4.70 |
ENSDART00000003242
|
zgc:123275
|
zgc:123275 |
| chr11_+_14321113 | 4.70 |
ENSDART00000039822
ENSDART00000137347 ENSDART00000132997 |
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr18_-_7031409 | 4.70 |
ENSDART00000148485
ENSDART00000005405 |
calub
|
calumenin b |
| chr6_+_612594 | 4.67 |
ENSDART00000150903
|
kynu
|
kynureninase |
| chr16_+_46410520 | 4.63 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr20_-_25486384 | 4.63 |
ENSDART00000141340
|
si:dkey-183n20.15
|
si:dkey-183n20.15 |
| chr6_-_50704689 | 4.59 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| chr13_+_46941930 | 4.58 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr10_-_4980150 | 4.58 |
ENSDART00000093228
|
mat2al
|
methionine adenosyltransferase II, alpha-like |
| chr18_+_20047374 | 4.58 |
ENSDART00000146957
|
uacaa
|
uveal autoantigen with coiled-coil domains and ankyrin repeats a |
| chr14_+_32942063 | 4.55 |
ENSDART00000187705
|
lnx2b
|
ligand of numb-protein X 2b |
| chr17_+_4030493 | 4.52 |
ENSDART00000151849
|
hao1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr21_-_45073 | 4.52 |
ENSDART00000185997
|
bhmt
|
betaine-homocysteine methyltransferase |
| chr22_-_4769140 | 4.49 |
ENSDART00000165235
|
calr3a
|
calreticulin 3a |
| chr11_+_14286160 | 4.47 |
ENSDART00000166236
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr4_+_13568469 | 4.45 |
ENSDART00000171235
ENSDART00000136152 |
calua
|
calumenin a |
| chr1_+_58840889 | 4.45 |
ENSDART00000098308
|
tmed1b
|
transmembrane p24 trafficking protein 1b |
| chr12_+_10706772 | 4.43 |
ENSDART00000158227
|
top2a
|
DNA topoisomerase II alpha |
| chr4_-_20043484 | 4.43 |
ENSDART00000167780
|
zgc:193726
|
zgc:193726 |
| chr6_-_9676108 | 4.42 |
ENSDART00000169915
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr16_+_43401005 | 4.40 |
ENSDART00000110994
|
sqlea
|
squalene epoxidase a |
| chr25_+_7492663 | 4.39 |
ENSDART00000166496
|
cat
|
catalase |
| chr7_+_57088920 | 4.39 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr2_+_49457626 | 4.38 |
ENSDART00000129967
|
sh3gl1a
|
SH3-domain GRB2-like 1a |
| chr11_-_37997419 | 4.33 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr3_-_62087346 | 4.33 |
ENSDART00000092665
|
srebf1
|
sterol regulatory element binding transcription factor 1 |
| chr4_+_9836465 | 4.32 |
ENSDART00000004879
|
hsp90b1
|
heat shock protein 90, beta (grp94), member 1 |
| chr16_-_31644545 | 4.32 |
ENSDART00000181634
|
CR855311.5
|
|
| chr17_+_23556764 | 4.31 |
ENSDART00000146787
|
pank1a
|
pantothenate kinase 1a |
| chr14_-_35892767 | 4.29 |
ENSDART00000052648
|
tmem144b
|
transmembrane protein 144b |
| chr9_-_1604601 | 4.26 |
ENSDART00000143130
|
agps
|
alkylglycerone phosphate synthase |
| chr15_+_14856307 | 4.26 |
ENSDART00000167213
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr8_-_40555340 | 4.26 |
ENSDART00000163348
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr6_-_53048291 | 4.25 |
ENSDART00000103267
|
fam212ab
|
family with sequence similarity 212, member Ab |
| chr20_-_29498178 | 4.24 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr12_+_47663419 | 4.24 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr24_-_36723116 | 4.23 |
ENSDART00000088204
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr3_+_7808459 | 4.22 |
ENSDART00000162374
|
hook2
|
hook microtubule-tethering protein 2 |
| chr10_+_29771256 | 4.22 |
ENSDART00000193195
|
hyou1
|
hypoxia up-regulated 1 |
| chr8_-_20230559 | 4.21 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr25_+_37446861 | 4.20 |
ENSDART00000189250
|
FO834799.1
|
|
| chr6_+_612330 | 4.17 |
ENSDART00000166872
ENSDART00000191758 |
kynu
|
kynureninase |
| chr3_+_48842918 | 4.17 |
ENSDART00000159420
|
hs3st3b1a
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1a |
| chr8_+_6576940 | 4.15 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr10_+_7703251 | 4.14 |
ENSDART00000165134
|
ggcx
|
gamma-glutamyl carboxylase |
| chr14_-_46897067 | 4.14 |
ENSDART00000058789
|
qdpra
|
quinoid dihydropteridine reductase a |
| chr22_-_35194187 | 4.14 |
ENSDART00000164443
ENSDART00000104687 |
pfn2
|
profilin 2 |
| chr7_-_48251234 | 4.14 |
ENSDART00000024062
ENSDART00000098904 |
cpeb1b
|
cytoplasmic polyadenylation element binding protein 1b |
| chr14_-_246342 | 4.14 |
ENSDART00000054823
|
aurkb
|
aurora kinase B |
| chr2_-_17115256 | 4.13 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr20_-_4793450 | 4.13 |
ENSDART00000053870
|
galca
|
galactosylceramidase a |
| chr9_+_21146862 | 4.13 |
ENSDART00000136365
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr6_-_39198912 | 4.11 |
ENSDART00000077938
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr1_-_54972170 | 4.10 |
ENSDART00000150548
ENSDART00000038330 |
khsrp
|
KH-type splicing regulatory protein |
| chr1_+_25696798 | 4.10 |
ENSDART00000054228
|
lrata
|
lecithin retinol acyltransferase a |
| chr21_-_32781612 | 4.10 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr12_-_17655683 | 4.06 |
ENSDART00000066411
|
dlgap5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr1_+_30723380 | 4.04 |
ENSDART00000127943
ENSDART00000062628 ENSDART00000127670 |
bora
|
bora, aurora kinase A activator |
| chr23_+_32028574 | 4.03 |
ENSDART00000145501
ENSDART00000143121 ENSDART00000111877 |
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr15_-_23793641 | 4.03 |
ENSDART00000122891
|
tmem97
|
transmembrane protein 97 |
| chr11_-_10456387 | 4.00 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr1_+_30723677 | 4.00 |
ENSDART00000177900
|
bora
|
bora, aurora kinase A activator |
| chr13_-_45063686 | 3.98 |
ENSDART00000130467
ENSDART00000136679 |
chp1
|
calcineurin-like EF-hand protein 1 |
| chr1_+_12348213 | 3.98 |
ENSDART00000144920
ENSDART00000138759 ENSDART00000067082 |
clta
|
clathrin, light chain A |
| chr14_-_25444774 | 3.98 |
ENSDART00000183448
|
slc26a2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr25_-_12809361 | 3.92 |
ENSDART00000162750
|
ca5a
|
carbonic anhydrase Va |
| chr4_-_4751981 | 3.90 |
ENSDART00000147436
ENSDART00000092984 ENSDART00000158466 |
creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr19_-_15420678 | 3.90 |
ENSDART00000151454
ENSDART00000027697 |
serinc2
|
serine incorporator 2 |
| chr12_+_5048044 | 3.90 |
ENSDART00000161548
ENSDART00000172607 |
kif22
|
kinesin family member 22 |
| chr16_-_12060488 | 3.89 |
ENSDART00000188733
|
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr3_+_13511984 | 3.85 |
ENSDART00000158942
|
CABZ01015475.1
|
|
| chr3_+_17951790 | 3.83 |
ENSDART00000164663
|
aclya
|
ATP citrate lyase a |
| chr21_+_10702031 | 3.82 |
ENSDART00000102304
|
lman1
|
lectin, mannose-binding, 1 |
| chr21_-_26406244 | 3.80 |
ENSDART00000137312
ENSDART00000077200 |
eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr8_+_16758304 | 3.80 |
ENSDART00000133514
|
elovl7a
|
ELOVL fatty acid elongase 7a |
| chr14_-_40821411 | 3.79 |
ENSDART00000166621
|
elf1
|
E74-like ETS transcription factor 1 |
| chr23_+_26733232 | 3.79 |
ENSDART00000035080
|
zgc:158263
|
zgc:158263 |
| chr18_+_45573416 | 3.78 |
ENSDART00000132184
ENSDART00000145288 |
kifc3
|
kinesin family member C3 |
| chr19_+_6990970 | 3.77 |
ENSDART00000158758
ENSDART00000160482 ENSDART00000193566 |
kifc1
|
kinesin family member C1 |
| chr5_-_54714789 | 3.77 |
ENSDART00000063357
|
ccnb1
|
cyclin B1 |
| chr25_+_37443194 | 3.76 |
ENSDART00000163178
ENSDART00000190262 |
slc10a3
|
solute carrier family 10, member 3 |
| chr6_+_41038757 | 3.76 |
ENSDART00000011245
|
entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr18_+_17600570 | 3.75 |
ENSDART00000175258
ENSDART00000151850 ENSDART00000151934 |
herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr2_+_49457449 | 3.72 |
ENSDART00000185470
|
sh3gl1a
|
SH3-domain GRB2-like 1a |
| chr6_-_39199070 | 3.72 |
ENSDART00000131793
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr10_-_39283883 | 3.72 |
ENSDART00000023831
|
cry5
|
cryptochrome circadian clock 5 |
| chr21_+_244503 | 3.71 |
ENSDART00000162889
|
stard4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr19_+_7636941 | 3.70 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr3_-_70782 | 3.67 |
ENSDART00000110602
|
zgc:165518
|
zgc:165518 |
Network of associatons between targets according to the STRING database.
First level regulatory network of fosaa+fosab+fosl1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.0 | GO:1902767 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 2.9 | 8.8 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 2.9 | 2.9 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 2.7 | 8.2 | GO:0006565 | L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 2.6 | 10.5 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 2.3 | 11.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 2.3 | 6.8 | GO:0036315 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 2.2 | 6.7 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 2.2 | 8.9 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 1.9 | 9.5 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 1.9 | 46.8 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 1.9 | 7.4 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 1.7 | 5.0 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 1.6 | 33.9 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.6 | 4.7 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 1.5 | 3.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 1.5 | 13.7 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 1.5 | 8.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 1.4 | 5.8 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 1.4 | 18.5 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 1.4 | 5.6 | GO:0052746 | inositol phosphorylation(GO:0052746) |
| 1.4 | 5.6 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 1.4 | 4.1 | GO:0001783 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 1.3 | 4.0 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 1.3 | 3.9 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 1.2 | 3.7 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 1.2 | 4.9 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 1.2 | 6.0 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 1.2 | 3.5 | GO:0051228 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 1.1 | 5.7 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 1.1 | 4.6 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 1.1 | 3.4 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 1.1 | 5.6 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 1.1 | 2.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 1.1 | 4.3 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 1.1 | 5.3 | GO:0070814 | cysteine biosynthetic process from serine(GO:0006535) hydrogen sulfide biosynthetic process(GO:0070814) |
| 1.1 | 3.2 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 1.0 | 3.1 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 1.0 | 3.0 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 1.0 | 12.1 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 1.0 | 9.8 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 1.0 | 6.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.0 | 7.7 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.9 | 1.9 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.9 | 3.7 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.9 | 16.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.9 | 38.0 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.9 | 6.5 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.9 | 5.5 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.9 | 5.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.9 | 3.5 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.9 | 6.2 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.9 | 2.6 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.9 | 7.7 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.9 | 4.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.8 | 4.1 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.8 | 9.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.8 | 2.4 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.8 | 3.1 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.7 | 3.7 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.7 | 2.9 | GO:0060471 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.7 | 4.3 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.7 | 4.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.7 | 2.0 | GO:0010656 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.7 | 5.4 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.7 | 2.0 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.6 | 4.5 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.6 | 12.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.6 | 7.3 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.6 | 6.7 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.6 | 4.8 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.6 | 4.1 | GO:0006559 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.6 | 2.4 | GO:0001715 | ectodermal cell fate commitment(GO:0001712) ectodermal cell fate specification(GO:0001715) ectodermal cell differentiation(GO:0010668) regulation of ectodermal cell fate specification(GO:0042665) regulation of ectoderm development(GO:2000383) |
| 0.6 | 2.3 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.6 | 4.5 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.6 | 5.6 | GO:0071451 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.6 | 3.4 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.5 | 2.7 | GO:0070861 | regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.5 | 5.9 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.5 | 1.1 | GO:0002828 | regulation of type 2 immune response(GO:0002828) |
| 0.5 | 5.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.5 | 3.1 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.5 | 4.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.5 | 2.1 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.5 | 13.8 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.5 | 5.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.5 | 1.0 | GO:0035089 | establishment of apical/basal cell polarity(GO:0035089) establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.5 | 1.5 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.5 | 6.5 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.5 | 3.0 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.5 | 3.0 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.5 | 3.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.5 | 5.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.5 | 1.9 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.5 | 2.8 | GO:1904666 | regulation of ubiquitin protein ligase activity(GO:1904666) positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.5 | 1.9 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.5 | 2.8 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.5 | 1.9 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.5 | 0.9 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.5 | 13.2 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.5 | 1.4 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.4 | 2.2 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.4 | 1.8 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.4 | 4.3 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.4 | 0.9 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.4 | 2.1 | GO:0034475 | U4 snRNA 3'-end processing(GO:0034475) |
| 0.4 | 2.6 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.4 | 1.7 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.4 | 1.2 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.4 | 0.8 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.4 | 4.9 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.4 | 2.8 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.4 | 2.4 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.4 | 1.6 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.4 | 1.6 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.4 | 2.4 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.4 | 5.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.4 | 8.7 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.4 | 0.8 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.4 | 9.4 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.4 | 1.5 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.4 | 0.8 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.4 | 1.5 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.4 | 2.3 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.4 | 4.2 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.4 | 3.8 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.4 | 1.5 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.4 | 3.0 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.4 | 6.7 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.4 | 3.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.4 | 11.6 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.4 | 2.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.4 | 1.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.3 | 6.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.3 | 3.1 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.3 | 1.4 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.3 | 1.4 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.3 | 4.4 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.3 | 11.4 | GO:0001878 | response to yeast(GO:0001878) |
| 0.3 | 0.7 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.3 | 1.0 | GO:0050428 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.3 | 2.3 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.3 | 2.3 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.3 | 1.3 | GO:0070586 | cell-cell adhesion involved in gastrulation(GO:0070586) |
| 0.3 | 7.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.3 | 4.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.3 | 1.9 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.3 | 0.9 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.3 | 2.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.3 | 1.2 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.3 | 4.5 | GO:0003323 | type B pancreatic cell development(GO:0003323) methionine biosynthetic process(GO:0009086) |
| 0.3 | 0.9 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.3 | 0.6 | GO:0019344 | sulfur amino acid biosynthetic process(GO:0000097) cysteine metabolic process(GO:0006534) homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.3 | 1.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.3 | 3.3 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.3 | 1.8 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.3 | 2.1 | GO:0046292 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.3 | 2.6 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.3 | 0.9 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.3 | 2.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 2.3 | GO:0031577 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) negative regulation of metaphase/anaphase transition of cell cycle(GO:1902100) |
| 0.3 | 5.4 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.3 | 0.6 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.3 | 1.4 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.3 | 12.8 | GO:0031017 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.3 | 1.9 | GO:0030728 | ovulation(GO:0030728) |
| 0.3 | 1.6 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.3 | 1.4 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.3 | 4.8 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.3 | 1.1 | GO:1904589 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.3 | 4.7 | GO:0030168 | platelet activation(GO:0030168) |
| 0.3 | 1.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.3 | 3.7 | GO:0042026 | protein refolding(GO:0042026) |
| 0.3 | 0.8 | GO:0007063 | regulation of sister chromatid cohesion(GO:0007063) regulation of chromosome condensation(GO:0060623) regulation of cohesin loading(GO:0071922) |
| 0.3 | 1.0 | GO:0006901 | vesicle coating(GO:0006901) |
| 0.3 | 1.3 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.3 | 2.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.3 | 1.8 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.3 | 24.8 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.3 | 2.3 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.2 | 11.2 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.2 | 0.5 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.2 | 2.0 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.2 | 20.1 | GO:0034976 | response to endoplasmic reticulum stress(GO:0034976) |
| 0.2 | 1.0 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.2 | 6.6 | GO:0046463 | triglyceride biosynthetic process(GO:0019432) neutral lipid biosynthetic process(GO:0046460) acylglycerol biosynthetic process(GO:0046463) |
| 0.2 | 0.9 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.2 | 0.9 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.2 | 22.3 | GO:0006956 | complement activation(GO:0006956) |
| 0.2 | 0.9 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.2 | 1.1 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.2 | 0.7 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.2 | 0.9 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.2 | 10.6 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.2 | 0.6 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.2 | 6.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 3.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.2 | 2.5 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.2 | 5.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 1.4 | GO:1900117 | regulation of execution phase of apoptosis(GO:1900117) negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 1.0 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.2 | 7.9 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.2 | 5.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.2 | 2.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 1.6 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 2.0 | GO:0010890 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.2 | 3.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 1.0 | GO:0010990 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 1.7 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.2 | 2.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 2.8 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) mitotic DNA integrity checkpoint(GO:0044774) |
| 0.2 | 0.6 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.2 | 0.7 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 1.3 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.2 | 1.1 | GO:0036294 | cellular response to decreased oxygen levels(GO:0036294) cellular response to oxygen levels(GO:0071453) |
| 0.2 | 2.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 1.9 | GO:0048339 | paraxial mesoderm development(GO:0048339) |
| 0.2 | 1.1 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.2 | 0.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.2 | 0.9 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.2 | 1.7 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.2 | 7.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.2 | 1.6 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.2 | 1.3 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.2 | 1.7 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.2 | 2.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 3.6 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.2 | 1.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 3.9 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.2 | 0.6 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 9.4 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.2 | 0.8 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 1.3 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.1 | 6.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 1.0 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 3.7 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 1.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.7 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 1.9 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.9 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 1.4 | GO:0030719 | P granule organization(GO:0030719) |
| 0.1 | 1.1 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 6.1 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 2.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 2.3 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 2.7 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 1.7 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 7.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 1.5 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 2.9 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.1 | 6.2 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 2.5 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 1.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 1.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.7 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.9 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 1.7 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 1.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 7.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.3 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 3.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 19.1 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 1.7 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 0.7 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 1.1 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 0.8 | GO:0000423 | macromitophagy(GO:0000423) C-terminal protein lipidation(GO:0006501) |
| 0.1 | 8.0 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 1.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 2.6 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 1.4 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 1.8 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 1.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 1.1 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.2 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 1.4 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 | 1.6 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 | 0.1 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.1 | 5.3 | GO:0034249 | negative regulation of translation(GO:0017148) negative regulation of cellular amide metabolic process(GO:0034249) |
| 0.1 | 0.5 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.8 | GO:0018202 | peptidyl-lysine monomethylation(GO:0018026) peptidyl-histidine modification(GO:0018202) |
| 0.1 | 1.1 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 1.4 | GO:0048096 | positive regulation of gene expression, epigenetic(GO:0045815) chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.9 | GO:0007530 | sex determination(GO:0007530) |
| 0.1 | 1.6 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 2.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 1.8 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 0.8 | GO:0021707 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.3 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.1 | 6.3 | GO:0008202 | steroid metabolic process(GO:0008202) |
| 0.1 | 11.4 | GO:0051346 | negative regulation of hydrolase activity(GO:0051346) |
| 0.1 | 0.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 2.2 | GO:0043039 | amino acid activation(GO:0043038) tRNA aminoacylation(GO:0043039) |
| 0.1 | 3.0 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.8 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.4 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 7.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 0.8 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 2.9 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 1.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 2.2 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 3.5 | GO:0048477 | oogenesis(GO:0048477) |
| 0.1 | 0.4 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.1 | 1.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.4 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 3.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 5.1 | GO:0010942 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.1 | 1.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 2.7 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.1 | 3.9 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.1 | 0.4 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.1 | 1.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.9 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.1 | 0.9 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 2.9 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 0.5 | GO:0071623 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 1.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 1.5 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 1.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.7 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.7 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 2.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.4 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.1 | 1.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 4.2 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.1 | 0.5 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 1.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.2 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) V(D)J recombination(GO:0033151) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 2.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 0.2 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.1 | 1.5 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.1 | 2.9 | GO:0050673 | epithelial cell proliferation(GO:0050673) |
| 0.1 | 1.0 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 2.0 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 1.2 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 0.8 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 1.2 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 1.6 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.3 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.8 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 2.8 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 1.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.9 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 2.1 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.2 | GO:0061383 | trabecula morphogenesis(GO:0061383) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.3 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 2.6 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.0 | 0.7 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.0 | 6.5 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 1.6 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 6.0 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.5 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.9 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 1.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 1.2 | GO:0050657 | nucleic acid transport(GO:0050657) RNA transport(GO:0050658) establishment of RNA localization(GO:0051236) |
| 0.0 | 1.4 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.7 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.5 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 7.6 | GO:0006396 | RNA processing(GO:0006396) |
| 0.0 | 1.1 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.7 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 3.9 | GO:0010876 | lipid localization(GO:0010876) |
| 0.0 | 0.0 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.3 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.3 | GO:0014068 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 1.5 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.9 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.2 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.0 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.2 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 18.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 4.6 | 13.7 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 2.3 | 6.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 2.1 | 12.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 1.5 | 13.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 1.5 | 4.4 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 1.4 | 4.3 | GO:0098536 | deuterosome(GO:0098536) |
| 1.3 | 9.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 1.1 | 12.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 1.1 | 3.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 1.0 | 4.0 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 1.0 | 5.0 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.9 | 2.8 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.9 | 3.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.9 | 3.5 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.9 | 40.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.8 | 5.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.8 | 2.4 | GO:0030689 | Noc complex(GO:0030689) |
| 0.7 | 3.7 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.7 | 4.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.7 | 4.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.7 | 2.0 | GO:0034709 | methylosome(GO:0034709) |
| 0.7 | 2.6 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.6 | 2.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.6 | 2.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.6 | 11.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.6 | 2.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.5 | 68.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.5 | 3.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.5 | 1.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.4 | 1.2 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.4 | 1.2 | GO:0097361 | CIA complex(GO:0097361) |
| 0.3 | 5.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.3 | 2.4 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.3 | 4.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.3 | 13.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.3 | 1.6 | GO:1990923 | PET complex(GO:1990923) |
| 0.3 | 6.5 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.3 | 3.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.3 | 15.2 | GO:0005657 | replication fork(GO:0005657) |
| 0.3 | 9.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.3 | 12.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.3 | 1.1 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.3 | 4.0 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.3 | 18.0 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.3 | 5.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.3 | 2.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 3.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 0.5 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.2 | 2.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 26.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 0.9 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.2 | 1.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 3.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 1.6 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 10.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 2.1 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.2 | 1.7 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.2 | 1.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.2 | 100.9 | GO:0042175 | nuclear outer membrane-endoplasmic reticulum membrane network(GO:0042175) |
| 0.2 | 0.5 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.2 | 4.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 1.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 2.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 9.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 1.0 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 1.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 3.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.2 | 0.9 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 2.1 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.9 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 2.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 6.8 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 14.8 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 0.4 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 2.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 3.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 2.2 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 2.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 16.4 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 2.1 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) |
| 0.1 | 0.3 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.1 | 1.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 1.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.4 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 2.2 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 0.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 5.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 15.4 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 4.1 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 1.5 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.1 | 5.1 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.1 | 0.7 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 2.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 4.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 2.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 3.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.4 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 24.1 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.1 | 6.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 2.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.1 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.0 | 2.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.8 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 1.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 4.6 | GO:0044440 | endosomal part(GO:0044440) |
| 0.0 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 25.1 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 5.6 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 2.9 | GO:0005882 | intermediate filament(GO:0005882) intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 4.3 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.5 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 1.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 22.7 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 0.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.0 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 2.9 | 17.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 2.8 | 11.3 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 2.8 | 8.3 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 2.7 | 8.2 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 2.6 | 7.9 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 2.3 | 11.6 | GO:0098639 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 2.2 | 6.5 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 2.0 | 14.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 1.9 | 7.4 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 1.8 | 5.4 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 1.8 | 8.8 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 1.7 | 6.8 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 1.6 | 33.9 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 1.5 | 7.7 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 1.5 | 4.4 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 1.5 | 7.3 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 1.4 | 5.8 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 1.4 | 4.1 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 1.3 | 6.4 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 1.3 | 3.8 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 1.2 | 3.7 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 1.2 | 11.8 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 1.2 | 15.1 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 1.1 | 5.5 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 1.1 | 4.4 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 1.1 | 5.3 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 1.0 | 3.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 1.0 | 4.1 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 1.0 | 4.1 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 1.0 | 7.2 | GO:0035173 | histone kinase activity(GO:0035173) |
| 1.0 | 3.0 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 1.0 | 6.7 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.9 | 8.5 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.9 | 24.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.9 | 8.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.9 | 13.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.9 | 3.5 | GO:0045547 | dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.8 | 4.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.8 | 5.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.8 | 3.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.8 | 3.8 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.7 | 7.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.7 | 4.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.7 | 2.7 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.7 | 3.4 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.7 | 10.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.7 | 2.0 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.7 | 3.3 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.6 | 3.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.6 | 6.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.6 | 6.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.6 | 1.8 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.6 | 2.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.6 | 7.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.6 | 4.5 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.6 | 6.7 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.5 | 9.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.5 | 4.9 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.5 | 7.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.5 | 2.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.5 | 2.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.5 | 1.5 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.5 | 7.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.5 | 4.4 | GO:0004096 | catalase activity(GO:0004096) |
| 0.5 | 6.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.5 | 3.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.5 | 4.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.5 | 3.7 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.5 | 4.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.5 | 13.2 | GO:0050661 | NADP binding(GO:0050661) |
| 0.4 | 6.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.4 | 1.7 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.4 | 2.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.4 | 2.9 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.4 | 2.5 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.4 | 2.8 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.4 | 6.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.4 | 25.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.4 | 9.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.4 | 3.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.4 | 6.1 | GO:0022884 | macromolecule transmembrane transporter activity(GO:0022884) |
| 0.4 | 1.5 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.4 | 3.0 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.4 | 1.9 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.4 | 1.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.4 | 10.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.4 | 1.4 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.4 | 1.1 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.4 | 2.8 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.4 | 1.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.3 | 3.8 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.3 | 2.4 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.3 | 7.4 | GO:0010181 | FMN binding(GO:0010181) |
| 0.3 | 1.0 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.3 | 2.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.3 | 1.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.3 | 2.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.3 | 0.9 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.3 | 8.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 0.9 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.3 | 5.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 5.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.3 | 6.9 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.3 | 5.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.3 | 1.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.3 | 4.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 3.7 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.3 | 1.7 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.3 | 1.6 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.3 | 4.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 9.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.3 | 0.8 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.3 | 2.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.3 | 2.6 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.2 | 1.7 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 5.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 2.4 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.2 | 3.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.2 | 2.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.2 | 17.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 3.8 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 0.6 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.2 | 1.0 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.2 | 9.1 | GO:0005347 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.2 | 3.7 | GO:0004532 | exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.2 | 2.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 8.0 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.2 | 2.8 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 2.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 3.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.2 | 1.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 0.8 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 1.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 0.8 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.2 | 1.7 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 1.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.2 | 3.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 1.7 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.2 | 1.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 1.9 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 4.9 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.2 | 1.0 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 1.4 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.2 | 3.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 4.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 6.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 12.1 | GO:0051536 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.2 | 0.9 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 0.8 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.2 | 1.6 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 2.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 2.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 2.7 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 1.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 2.9 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 1.0 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 1.9 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 5.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 23.5 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 1.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.5 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 7.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.9 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 3.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 10.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.5 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 1.7 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 6.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.8 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 1.9 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 1.0 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 2.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 29.4 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 8.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 4.8 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 0.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.8 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 2.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 6.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 2.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 2.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 5.7 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 2.2 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 9.0 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.1 | 2.1 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 2.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.6 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 3.9 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 0.6 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 1.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.9 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 3.2 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.1 | 0.8 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 0.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 1.1 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.1 | 1.0 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 13.1 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 4.6 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 1.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.2 | GO:0032143 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.3 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 2.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 6.5 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 1.1 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 1.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.5 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 2.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 3.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 10.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 1.0 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.1 | 1.3 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.1 | 1.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.5 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 8.9 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 3.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 1.7 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.7 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 1.8 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.1 | 3.7 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.1 | 8.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.2 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 2.3 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 3.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.5 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 1.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.5 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.3 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 0.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 2.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.7 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 2.4 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.9 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 0.5 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 1.1 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 3.0 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 1.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 1.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 4.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 1.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.5 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 1.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.8 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 12.5 | GO:0016462 | pyrophosphatase activity(GO:0016462) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.9 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 2.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 7.9 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 1.3 | 61.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.7 | 23.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.6 | 9.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.6 | 22.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.6 | 22.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.4 | 4.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.4 | 11.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.4 | 4.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.3 | 9.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.3 | 6.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.3 | 2.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.3 | 7.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.3 | 1.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 8.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.3 | 4.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.3 | 4.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 7.4 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.2 | 2.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 13.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 6.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 10.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 5.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 2.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 4.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 0.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 1.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 2.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 0.8 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 3.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 3.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 2.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.2 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 2.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 1.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 2.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 0.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 2.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 0.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 0.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 80.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 3.3 | 32.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 3.2 | 9.7 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 2.8 | 2.8 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 2.1 | 25.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 2.1 | 18.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 1.7 | 12.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 1.4 | 26.7 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 1.2 | 13.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 1.1 | 23.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.9 | 8.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.9 | 4.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.9 | 16.2 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.8 | 18.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.8 | 4.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.8 | 3.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.7 | 12.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.7 | 9.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.7 | 4.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.6 | 7.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.6 | 16.1 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.6 | 7.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.5 | 11.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.5 | 4.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.4 | 6.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.4 | 20.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.4 | 17.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.4 | 14.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.4 | 6.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.3 | 5.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.3 | 7.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.3 | 1.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.3 | 19.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.3 | 1.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 1.8 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.2 | 1.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 5.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 1.4 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.2 | 6.2 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.2 | 3.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 4.9 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.2 | 8.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 1.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 9.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 4.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.2 | 0.8 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.2 | 1.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 2.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 3.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 2.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 1.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 1.5 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.2 | 3.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 1.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 8.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.2 | 0.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 0.9 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.2 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 2.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 3.5 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 1.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 0.9 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 3.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 0.5 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.1 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.1 | 1.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 0.8 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 10.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 3.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 11.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 2.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 3.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 11.6 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 1.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.3 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.1 | 4.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 3.2 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 1.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.9 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 1.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 2.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.5 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 1.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.5 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 4.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.4 | REACTOME DNA REPAIR | Genes involved in DNA Repair |