Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
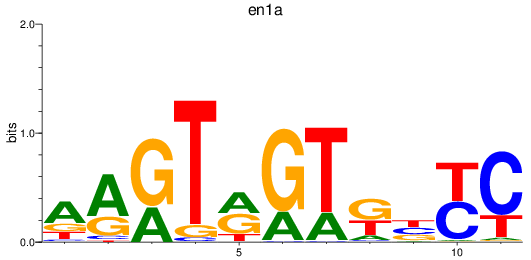
Results for en1a
Z-value: 1.29
Transcription factors associated with en1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
en1a
|
ENSDARG00000014321 | engrailed homeobox 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| en1a | dr11_v1_chr9_-_785444_785444 | -0.63 | 6.5e-12 | Click! |
Activity profile of en1a motif
Sorted Z-values of en1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_-_26018424 | 19.78 |
ENSDART00000089332
|
acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr2_-_51794472 | 17.87 |
ENSDART00000186652
|
BX908782.3
|
|
| chr16_+_23924040 | 17.71 |
ENSDART00000161124
|
si:dkey-7f3.14
|
si:dkey-7f3.14 |
| chr24_-_24983047 | 16.49 |
ENSDART00000066631
|
slc51a
|
solute carrier family 51, alpha subunit |
| chr10_+_28428222 | 16.40 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr25_-_26018240 | 16.16 |
ENSDART00000150800
|
acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr8_-_50147948 | 15.87 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr20_-_25518488 | 14.40 |
ENSDART00000186993
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr13_-_34858500 | 12.98 |
ENSDART00000184843
|
sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr14_-_21123551 | 12.48 |
ENSDART00000171679
ENSDART00000165882 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr23_+_28770225 | 12.13 |
ENSDART00000132179
ENSDART00000142273 |
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr13_-_15700060 | 12.08 |
ENSDART00000170689
ENSDART00000010986 ENSDART00000101741 ENSDART00000139124 |
ckba
|
creatine kinase, brain a |
| chr2_-_1514001 | 11.58 |
ENSDART00000057736
|
c8b
|
complement component 8, beta polypeptide |
| chr20_+_43942278 | 11.48 |
ENSDART00000100571
|
clic5b
|
chloride intracellular channel 5b |
| chr15_+_14854666 | 11.31 |
ENSDART00000163066
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr9_-_9998087 | 11.28 |
ENSDART00000124423
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr3_+_12732382 | 10.82 |
ENSDART00000158403
|
cyp2k19
|
cytochrome P450, family 2, subfamily k, polypeptide 19 |
| chr25_-_13023569 | 10.11 |
ENSDART00000167232
|
ccl39.1
|
chemokine (C-C motif) ligand 39, duplicate 1 |
| chr3_-_3939785 | 9.92 |
ENSDART00000049593
|
unm_sa1506
|
un-named sa1506 |
| chr11_-_669558 | 9.10 |
ENSDART00000173450
|
pparg
|
peroxisome proliferator-activated receptor gamma |
| chr20_+_34845672 | 9.07 |
ENSDART00000128895
|
emilin1a
|
elastin microfibril interfacer 1a |
| chr7_+_30178453 | 8.73 |
ENSDART00000174420
|
si:ch211-107o10.3
|
si:ch211-107o10.3 |
| chr10_+_38708099 | 8.41 |
ENSDART00000172306
|
tmprss2
|
transmembrane protease, serine 2 |
| chr7_+_49664174 | 8.18 |
ENSDART00000137059
ENSDART00000131210 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr7_+_44713135 | 8.18 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr16_+_26017360 | 8.09 |
ENSDART00000149466
|
prss59.2
|
protease, serine, 59, tandem duplicate 2 |
| chr3_+_26345732 | 7.87 |
ENSDART00000128613
|
rps15a
|
ribosomal protein S15a |
| chr11_-_669270 | 7.71 |
ENSDART00000172834
|
pparg
|
peroxisome proliferator-activated receptor gamma |
| chr5_-_44843738 | 7.25 |
ENSDART00000003926
|
fbp1a
|
fructose-1,6-bisphosphatase 1a |
| chr7_-_10560964 | 6.97 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr7_-_20582842 | 6.86 |
ENSDART00000169750
ENSDART00000111719 |
si:dkey-19b23.11
|
si:dkey-19b23.11 |
| chr2_+_26498446 | 6.15 |
ENSDART00000078412
|
rps8a
|
ribosomal protein S8a |
| chr12_-_26537145 | 6.10 |
ENSDART00000138437
ENSDART00000163931 ENSDART00000132737 |
acsf2
|
acyl-CoA synthetase family member 2 |
| chr5_+_22459087 | 5.99 |
ENSDART00000134781
|
BX546499.1
|
|
| chr16_+_23383316 | 5.96 |
ENSDART00000103220
|
krtcap2
|
keratinocyte associated protein 2 |
| chr25_-_30357027 | 5.88 |
ENSDART00000171137
|
pdia3
|
protein disulfide isomerase family A, member 3 |
| chr18_-_14901437 | 5.78 |
ENSDART00000145842
ENSDART00000008035 |
trabd
|
TraB domain containing |
| chr9_+_21277846 | 5.73 |
ENSDART00000139620
ENSDART00000110996 ENSDART00000111899 |
lats2
|
large tumor suppressor kinase 2 |
| chr5_-_8164439 | 5.61 |
ENSDART00000189912
|
slc1a3a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3a |
| chr17_+_3128273 | 5.37 |
ENSDART00000122453
|
zgc:136872
|
zgc:136872 |
| chr5_-_30615901 | 5.10 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr19_+_43780970 | 5.03 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr14_-_38872536 | 4.91 |
ENSDART00000184315
ENSDART00000127479 |
gsr
|
glutathione reductase |
| chr9_+_3113211 | 4.89 |
ENSDART00000139476
|
ppp1r9ala
|
protein phosphatase 1 regulatory subunit 9A-like A |
| chr20_+_36629173 | 4.86 |
ENSDART00000161241
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr6_+_23122789 | 4.83 |
ENSDART00000049226
ENSDART00000067560 |
acox1
|
acyl-CoA oxidase 1, palmitoyl |
| chr6_+_46320693 | 4.82 |
ENSDART00000128621
ENSDART00000155917 |
si:dkeyp-67f1.2
|
si:dkeyp-67f1.2 |
| chr9_+_23003208 | 4.80 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr23_-_33738945 | 4.79 |
ENSDART00000136386
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr22_-_31020690 | 4.79 |
ENSDART00000130604
|
ssuh2.4
|
ssu-2 homolog, tandem duplicate 4 |
| chr17_-_6076266 | 4.72 |
ENSDART00000171084
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr12_-_8070969 | 4.64 |
ENSDART00000020995
|
tmem26b
|
transmembrane protein 26b |
| chr4_+_28997595 | 4.62 |
ENSDART00000133357
|
si:dkey-13e3.1
|
si:dkey-13e3.1 |
| chr17_-_8886735 | 4.51 |
ENSDART00000121997
|
nkl.3
|
NK-lysin tandem duplicate 3 |
| chr13_+_15190677 | 4.38 |
ENSDART00000142240
ENSDART00000129045 |
mavs
|
mitochondrial antiviral signaling protein |
| chr14_-_11529311 | 4.36 |
ENSDART00000127208
|
si:ch211-153b23.7
|
si:ch211-153b23.7 |
| chr5_+_9360394 | 4.34 |
ENSDART00000124642
|
FP236810.2
|
|
| chr16_-_21489514 | 4.29 |
ENSDART00000149525
ENSDART00000148517 ENSDART00000146914 ENSDART00000186493 ENSDART00000193081 ENSDART00000186017 |
mpp6a
|
membrane protein, palmitoylated 6a (MAGUK p55 subfamily member 6) |
| chr14_+_45732081 | 4.28 |
ENSDART00000110103
|
ccdc88b
|
coiled-coil domain containing 88B |
| chr4_-_52165969 | 4.22 |
ENSDART00000171130
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr2_-_24398324 | 4.20 |
ENSDART00000165226
|
zgc:154006
|
zgc:154006 |
| chr23_-_10722664 | 4.16 |
ENSDART00000146526
ENSDART00000129022 ENSDART00000104985 |
foxp1a
|
forkhead box P1a |
| chr13_+_11876437 | 4.13 |
ENSDART00000179753
|
trim8a
|
tripartite motif containing 8a |
| chr17_-_6349044 | 4.07 |
ENSDART00000081707
|
oct2
|
organic cation transporter 2 |
| chr3_+_12784460 | 4.05 |
ENSDART00000168382
|
cyp2k8
|
cytochrome P450, family 2, subfamily K, polypeptide 8 |
| chr19_-_32914227 | 4.05 |
ENSDART00000186115
ENSDART00000124246 |
mtdha
|
metadherin a |
| chr1_-_11607062 | 3.95 |
ENSDART00000158253
|
si:dkey-26i13.6
|
si:dkey-26i13.6 |
| chr12_-_30540699 | 3.89 |
ENSDART00000167712
ENSDART00000102464 |
zgc:153920
|
zgc:153920 |
| chr5_-_69716501 | 3.88 |
ENSDART00000158956
|
mob1a
|
MOB kinase activator 1A |
| chr16_-_5873866 | 3.85 |
ENSDART00000136936
|
trak1a
|
trafficking protein, kinesin binding 1a |
| chr18_-_43866526 | 3.84 |
ENSDART00000111309
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr15_+_15779184 | 3.81 |
ENSDART00000156902
|
si:ch211-33e4.2
|
si:ch211-33e4.2 |
| chr14_+_9432627 | 3.78 |
ENSDART00000042727
|
si:dkeyp-86f7.4
|
si:dkeyp-86f7.4 |
| chr1_+_58205904 | 3.69 |
ENSDART00000147324
|
si:dkey-222h21.12
|
si:dkey-222h21.12 |
| chr24_-_2423791 | 3.67 |
ENSDART00000190402
|
rreb1a
|
ras responsive element binding protein 1a |
| chr5_-_65021736 | 3.66 |
ENSDART00000162368
ENSDART00000161876 |
anxa1c
|
annexin A1c |
| chr2_+_38025260 | 3.65 |
ENSDART00000075905
|
hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr4_+_66853199 | 3.60 |
ENSDART00000170223
|
si:dkey-29m1.2
|
si:dkey-29m1.2 |
| chr2_+_49631850 | 3.57 |
ENSDART00000114274
ENSDART00000114516 |
si:dkey-53k12.18
|
si:dkey-53k12.18 |
| chr2_+_45441563 | 3.57 |
ENSDART00000151882
|
si:ch211-66k16.27
|
si:ch211-66k16.27 |
| chr1_+_22654875 | 3.54 |
ENSDART00000019698
ENSDART00000161874 |
anxa5b
|
annexin A5b |
| chr5_-_38197080 | 3.51 |
ENSDART00000140708
|
si:ch211-284e13.9
|
si:ch211-284e13.9 |
| chr19_-_18626515 | 3.50 |
ENSDART00000160624
|
rps18
|
ribosomal protein S18 |
| chr11_-_3907937 | 3.47 |
ENSDART00000082409
|
rpn1
|
ribophorin I |
| chr16_-_46578523 | 3.47 |
ENSDART00000131061
|
si:dkey-152b24.6
|
si:dkey-152b24.6 |
| chr3_-_4557653 | 3.45 |
ENSDART00000111305
|
ftr50
|
finTRIM family, member 50 |
| chr6_+_6822592 | 3.41 |
ENSDART00000151057
|
si:ch211-85n16.4
|
si:ch211-85n16.4 |
| chr2_+_36114194 | 3.39 |
ENSDART00000113547
|
traj39
|
T-cell receptor alpha joining 39 |
| chr23_+_42819221 | 3.39 |
ENSDART00000180495
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr23_+_44374041 | 3.34 |
ENSDART00000136056
|
ephb4b
|
eph receptor B4b |
| chr9_+_38408991 | 3.32 |
ENSDART00000045588
|
cyp27a7
|
cytochrome P450, family 27, subfamily A, polypeptide 7 |
| chr17_-_23631400 | 3.32 |
ENSDART00000079563
|
fas
|
Fas cell surface death receptor |
| chr7_+_38507006 | 3.27 |
ENSDART00000173830
|
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr2_+_30969029 | 3.22 |
ENSDART00000085242
|
lpin2
|
lipin 2 |
| chr3_-_4579214 | 3.16 |
ENSDART00000164456
|
ftr50
|
finTRIM family, member 50 |
| chr17_-_6076084 | 3.16 |
ENSDART00000058890
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr8_+_25616946 | 3.15 |
ENSDART00000133983
|
slc38a5a
|
solute carrier family 38, member 5a |
| chr9_-_17417628 | 3.13 |
ENSDART00000060425
ENSDART00000141997 |
ftr53
|
finTRIM family, member 53 |
| chr1_-_48922273 | 3.11 |
ENSDART00000150254
|
si:ch211-112b1.2
|
si:ch211-112b1.2 |
| chr9_+_44994214 | 3.09 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr7_-_34265481 | 3.08 |
ENSDART00000173596
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr7_-_60096318 | 2.97 |
ENSDART00000189125
|
BX511067.1
|
|
| chr7_+_49695904 | 2.97 |
ENSDART00000183550
ENSDART00000126991 |
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr17_+_42274825 | 2.92 |
ENSDART00000020156
|
pax1a
|
paired box 1a |
| chr19_+_4076259 | 2.92 |
ENSDART00000160633
|
zgc:173581
|
zgc:173581 |
| chr4_-_17805128 | 2.90 |
ENSDART00000128988
|
spi2
|
Spi-2 proto-oncogene |
| chr2_+_22495274 | 2.85 |
ENSDART00000167915
|
lrrc8da
|
leucine rich repeat containing 8 VRAC subunit Da |
| chr3_-_32934359 | 2.80 |
ENSDART00000124160
|
aoc2
|
amine oxidase, copper containing 2 |
| chr15_-_20233105 | 2.74 |
ENSDART00000123910
|
ppp1r14ab
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ab |
| chr1_-_55131439 | 2.72 |
ENSDART00000074083
|
rab1aa
|
RAB1A, member RAS oncogene family a |
| chr12_-_44018667 | 2.70 |
ENSDART00000170692
|
si:dkey-201i2.4
|
si:dkey-201i2.4 |
| chr2_+_36606019 | 2.70 |
ENSDART00000098415
|
ing5b
|
inhibitor of growth family, member 5b |
| chr1_-_11606903 | 2.67 |
ENSDART00000136093
|
si:dkey-26i13.6
|
si:dkey-26i13.6 |
| chr18_-_17513426 | 2.66 |
ENSDART00000146725
|
b3gnt9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr8_-_38403018 | 2.65 |
ENSDART00000134100
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr18_-_7422787 | 2.58 |
ENSDART00000081386
|
si:dkey-30c15.12
|
si:dkey-30c15.12 |
| chr12_+_17754859 | 2.55 |
ENSDART00000112119
|
bhlha15
|
basic helix-loop-helix family, member a15 |
| chr23_-_28120058 | 2.50 |
ENSDART00000087815
|
b4galnt1a
|
beta-1,4-N-acetyl-galactosaminyl transferase 1a |
| chr9_+_55857193 | 2.50 |
ENSDART00000160980
|
sept10
|
septin 10 |
| chr22_+_19220459 | 2.49 |
ENSDART00000163070
|
si:dkey-21e2.7
|
si:dkey-21e2.7 |
| chr2_+_45479841 | 2.42 |
ENSDART00000151856
|
si:ch211-66k16.28
|
si:ch211-66k16.28 |
| chr17_-_45734459 | 2.42 |
ENSDART00000137807
|
arf6b
|
ADP-ribosylation factor 6b |
| chr13_-_1130096 | 2.41 |
ENSDART00000010261
|
pno1
|
partner of NOB1 homolog |
| chr8_-_1267247 | 2.40 |
ENSDART00000150064
|
cdc14b
|
cell division cycle 14B |
| chr2_-_3045861 | 2.39 |
ENSDART00000105818
ENSDART00000187575 |
guk1a
|
guanylate kinase 1a |
| chr7_-_24373662 | 2.39 |
ENSDART00000173865
|
PTGR1 (1 of many)
|
si:dkey-11k2.7 |
| chr17_-_45734231 | 2.36 |
ENSDART00000074873
|
arf6b
|
ADP-ribosylation factor 6b |
| chr11_+_24716837 | 2.35 |
ENSDART00000145217
|
zgc:153953
|
zgc:153953 |
| chr10_-_21542702 | 2.33 |
ENSDART00000146761
ENSDART00000134502 |
zgc:165539
|
zgc:165539 |
| chr23_-_20363261 | 2.33 |
ENSDART00000054651
|
si:rp71-17i16.5
|
si:rp71-17i16.5 |
| chr22_+_1123873 | 2.32 |
ENSDART00000137708
|
si:ch1073-181h11.2
|
si:ch1073-181h11.2 |
| chr8_+_40284973 | 2.30 |
ENSDART00000018401
|
orai1a
|
ORAI calcium release-activated calcium modulator 1a |
| chr2_+_32780138 | 2.28 |
ENSDART00000082250
|
zgc:136930
|
zgc:136930 |
| chr3_-_13921173 | 2.26 |
ENSDART00000159177
ENSDART00000165174 |
si:dkey-61n16.5
|
si:dkey-61n16.5 |
| chr20_+_51833030 | 2.26 |
ENSDART00000074330
|
taf1a
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A |
| chr22_-_23228669 | 2.25 |
ENSDART00000079536
|
nek7
|
NIMA-related kinase 7 |
| chr4_-_75707991 | 2.24 |
ENSDART00000166358
ENSDART00000160021 |
si:dkey-165o8.2
|
si:dkey-165o8.2 |
| chr9_+_38420028 | 2.21 |
ENSDART00000135748
|
cyp27a1.2
|
cytochrome P450, family 27, subfamily A, polypeptide 1, gene 2 |
| chr3_+_23768898 | 2.20 |
ENSDART00000110682
|
hoxb1a
|
homeobox B1a |
| chr8_+_43852743 | 2.18 |
ENSDART00000186485
|
AL808129.2
|
|
| chr1_+_12009673 | 2.18 |
ENSDART00000080100
|
slc24a2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr3_+_14543953 | 2.18 |
ENSDART00000161710
|
epor
|
erythropoietin receptor |
| chr13_+_18311410 | 2.17 |
ENSDART00000036718
ENSDART00000132073 |
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr4_+_57099307 | 2.17 |
ENSDART00000131654
|
si:ch211-238e22.2
|
si:ch211-238e22.2 |
| chr9_+_1313418 | 2.17 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr2_-_6518785 | 2.15 |
ENSDART00000146108
|
rgs1
|
regulator of G protein signaling 1 |
| chr22_-_37738203 | 2.12 |
ENSDART00000143190
|
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr6_+_58725653 | 2.12 |
ENSDART00000190584
|
igsf8
|
immunoglobulin superfamily, member 8 |
| chr5_-_57723929 | 2.08 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr16_-_41535690 | 2.06 |
ENSDART00000102662
|
rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| chr12_+_9342502 | 2.05 |
ENSDART00000152808
|
kcnh6b
|
potassium voltage-gated channel, subfamily H (eag-related), member 6b |
| chr1_-_11627289 | 2.05 |
ENSDART00000163312
|
si:dkey-26i13.3
|
si:dkey-26i13.3 |
| chr18_+_41232719 | 2.05 |
ENSDART00000138552
ENSDART00000145863 |
trip12
|
thyroid hormone receptor interactor 12 |
| chr11_-_13611132 | 1.95 |
ENSDART00000113055
|
si:ch211-1a19.2
|
si:ch211-1a19.2 |
| chr20_-_27152495 | 1.94 |
ENSDART00000020710
|
btbd7
|
BTB (POZ) domain containing 7 |
| chr6_+_21684296 | 1.90 |
ENSDART00000057223
|
rhebl1
|
Ras homolog, mTORC1 binding like 1 |
| chr2_-_6519017 | 1.87 |
ENSDART00000181716
|
rgs1
|
regulator of G protein signaling 1 |
| chr1_+_2301961 | 1.87 |
ENSDART00000108919
ENSDART00000143361 ENSDART00000142944 |
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr12_+_1385192 | 1.87 |
ENSDART00000160788
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr18_+_19456648 | 1.85 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr7_-_39751540 | 1.84 |
ENSDART00000016803
|
grpel1
|
GrpE-like 1, mitochondrial |
| chr7_+_17120811 | 1.83 |
ENSDART00000147140
|
prmt3
|
protein arginine methyltransferase 3 |
| chr11_+_13176568 | 1.83 |
ENSDART00000125371
ENSDART00000123257 |
mknk1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr20_-_20312789 | 1.83 |
ENSDART00000114779
|
si:ch211-212g7.6
|
si:ch211-212g7.6 |
| chr25_-_3836597 | 1.82 |
ENSDART00000115154
|
si:ch211-247i17.1
|
si:ch211-247i17.1 |
| chr10_+_38512270 | 1.82 |
ENSDART00000109752
|
serpinh1a
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1a |
| chr9_-_30555725 | 1.82 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr1_-_18585046 | 1.79 |
ENSDART00000147228
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr23_+_1730663 | 1.76 |
ENSDART00000149545
|
tgm1
|
transglutaminase 1, K polypeptide |
| chr14_+_26247319 | 1.75 |
ENSDART00000192793
|
CCDC69
|
coiled-coil domain containing 69 |
| chr15_-_1485086 | 1.75 |
ENSDART00000191651
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr2_-_26590628 | 1.74 |
ENSDART00000025120
|
ndc1
|
NDC1 transmembrane nucleoporin |
| chr21_-_21570134 | 1.71 |
ENSDART00000174388
|
or133-4
|
odorant receptor, family H, subfamily 133, member 4 |
| chr10_-_16062565 | 1.69 |
ENSDART00000188728
|
si:dkey-184a18.5
|
si:dkey-184a18.5 |
| chr23_-_26120281 | 1.68 |
ENSDART00000133372
ENSDART00000103872 |
rca2.1
|
regulator of complement activation group 2 gene 1 |
| chr19_-_29832876 | 1.67 |
ENSDART00000005119
|
eif3i
|
eukaryotic translation initiation factor 3, subunit I |
| chr7_-_17297156 | 1.67 |
ENSDART00000161336
|
nitr11a
|
novel immune-type receptor 11a |
| chr3_+_16663373 | 1.67 |
ENSDART00000100961
|
zgc:55558
|
zgc:55558 |
| chr17_-_465285 | 1.66 |
ENSDART00000168718
|
chrm5a
|
cholinergic receptor, muscarinic 5a |
| chr22_+_25733817 | 1.65 |
ENSDART00000176962
ENSDART00000187865 |
si:dkeyp-98a7.9
|
si:dkeyp-98a7.9 |
| chr7_+_31879986 | 1.64 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr3_-_26184018 | 1.64 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr15_+_25158104 | 1.62 |
ENSDART00000128267
|
slc35f2l
|
info solute carrier family 35, member F2, like |
| chr18_+_33322515 | 1.62 |
ENSDART00000136603
|
v2ra18
|
vomeronasal 2 receptor, a18 |
| chr4_+_77905736 | 1.60 |
ENSDART00000144471
|
si:zfos-2131b9.2
|
si:zfos-2131b9.2 |
| chr2_-_985417 | 1.60 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr2_-_3616363 | 1.60 |
ENSDART00000144699
|
pter
|
phosphotriesterase related |
| chr11_-_6880725 | 1.59 |
ENSDART00000007204
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr20_-_37813863 | 1.58 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr6_-_2134581 | 1.57 |
ENSDART00000175478
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr12_-_30523216 | 1.56 |
ENSDART00000152896
ENSDART00000153191 |
plekhs1
|
pleckstrin homology domain containing, family S member 1 |
| chr7_+_53199763 | 1.55 |
ENSDART00000160097
|
cdh28
|
cadherin 28 |
| chr7_-_73856073 | 1.55 |
ENSDART00000181793
|
FP236812.2
|
|
| chr5_-_68022631 | 1.54 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr12_-_10381282 | 1.52 |
ENSDART00000152788
|
mki67
|
marker of proliferation Ki-67 |
| chr10_-_39052264 | 1.51 |
ENSDART00000144036
|
igsf5a
|
immunoglobulin superfamily, member 5a |
| chr25_+_20140926 | 1.51 |
ENSDART00000121989
|
cald1b
|
caldesmon 1b |
| chr22_+_835728 | 1.51 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr25_+_19095231 | 1.49 |
ENSDART00000154066
|
isg20
|
interferon stimulated exonuclease gene |
| chr10_-_40429747 | 1.48 |
ENSDART00000150613
|
taar20r
|
trace amine associated receptor 20r |
Network of associatons between targets according to the STRING database.
First level regulatory network of en1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.2 | GO:0005986 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 2.3 | 11.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 2.0 | 6.0 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 1.8 | 5.5 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D biosynthetic process(GO:0042368) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 1.6 | 35.9 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 1.6 | 4.8 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 1.3 | 13.0 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 1.3 | 3.8 | GO:0005991 | disaccharide metabolic process(GO:0005984) trehalose metabolic process(GO:0005991) |
| 1.2 | 4.8 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 1.2 | 16.8 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 1.2 | 16.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 1.0 | 7.0 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.9 | 12.1 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.9 | 1.8 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.9 | 4.4 | GO:0039528 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.8 | 3.3 | GO:0070227 | lymphocyte apoptotic process(GO:0070227) |
| 0.8 | 3.2 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.8 | 12.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.7 | 2.2 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.6 | 3.9 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.6 | 3.7 | GO:1900138 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) negative regulation of phospholipase activity(GO:0010519) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) regulation of leukocyte apoptotic process(GO:2000106) positive regulation of leukocyte apoptotic process(GO:2000108) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.6 | 29.2 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.5 | 5.7 | GO:0046620 | secondary heart field specification(GO:0003139) regulation of organ growth(GO:0046620) |
| 0.5 | 3.7 | GO:0031179 | peptide modification(GO:0031179) |
| 0.5 | 1.4 | GO:0090266 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.4 | 3.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.4 | 12.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.4 | 2.0 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.4 | 7.9 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.4 | 3.5 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.3 | 2.0 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.3 | 2.2 | GO:0021661 | rhombomere 4 development(GO:0021570) rhombomere 4 morphogenesis(GO:0021661) |
| 0.3 | 4.8 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.3 | 2.7 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.2 | 2.4 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.2 | 10.1 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.2 | 1.9 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.2 | 2.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 0.7 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 5.0 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.2 | 1.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.2 | 1.9 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.2 | 6.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.2 | 1.0 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.2 | 3.5 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.2 | 2.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 2.7 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 4.9 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.2 | 4.8 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.2 | 1.4 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.2 | 4.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.2 | 4.9 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.2 | 1.7 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 1.7 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 2.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 2.7 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 0.8 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 2.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 1.8 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 0.7 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 1.5 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.7 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.7 | GO:0050927 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.1 | 1.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 3.6 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.2 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.1 | 0.7 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 2.3 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.1 | 0.3 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 3.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.5 | GO:2000290 | regulation of myotome development(GO:2000290) |
| 0.1 | 6.4 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 0.8 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.5 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 1.7 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 3.4 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.1 | 0.4 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.1 | 1.3 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 1.8 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 0.3 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 3.0 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.1 | 1.6 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.9 | GO:0061138 | morphogenesis of a branching epithelium(GO:0061138) |
| 0.1 | 8.2 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.1 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.7 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 1.1 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 1.9 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.4 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.0 | 3.2 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 1.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 2.5 | GO:0031017 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 0.4 | GO:1900186 | optomotor response(GO:0071632) caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 1.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 2.1 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 1.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 4.1 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.8 | GO:0031936 | nucleosome positioning(GO:0016584) negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.5 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.2 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 5.9 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 5.0 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 3.1 | GO:0009308 | amine metabolic process(GO:0009308) |
| 0.0 | 1.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.5 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 1.5 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.8 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 1.2 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 2.5 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 2.2 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 1.2 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.7 | GO:0001757 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.0 | 2.4 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 3.6 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 4.5 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.0 | 6.5 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 1.1 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 1.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 31.6 | GO:0006508 | proteolysis(GO:0006508) |
| 0.0 | 1.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 3.0 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
| 0.0 | 0.6 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.1 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 1.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 2.2 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 0.6 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 4.6 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.8 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 1.6 | GO:0006364 | rRNA processing(GO:0006364) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 13.0 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.8 | 2.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.7 | 11.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.6 | 4.8 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.5 | 1.8 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.5 | 1.8 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.4 | 1.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.3 | 2.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.3 | 14.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.3 | 1.8 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 3.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 3.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 2.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 1.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.2 | 1.0 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.2 | 0.7 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 0.5 | GO:0031213 | RSF complex(GO:0031213) |
| 0.2 | 0.9 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 2.7 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 3.5 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.1 | 12.7 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 2.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 5.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 5.0 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.1 | 7.3 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 1.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 4.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 2.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 2.5 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 2.6 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 0.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 44.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 5.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 3.6 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.0 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 2.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 2.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.5 | GO:0030425 | dendrite(GO:0030425) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 16.8 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 2.8 | 35.9 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 2.6 | 7.9 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 2.4 | 7.2 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 2.0 | 6.1 | GO:0047760 | medium-chain fatty acid-CoA ligase activity(GO:0031956) butyrate-CoA ligase activity(GO:0047760) |
| 1.9 | 13.0 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 1.8 | 5.5 | GO:0031073 | vitamin D3 25-hydroxylase activity(GO:0030343) cholesterol 26-hydroxylase activity(GO:0031073) |
| 1.6 | 4.9 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 1.3 | 3.8 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 1.0 | 4.9 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 1.0 | 4.8 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.9 | 12.1 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.8 | 4.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.7 | 3.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.6 | 3.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.6 | 10.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.6 | 2.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.5 | 2.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.5 | 1.8 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.5 | 5.9 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.4 | 29.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.4 | 3.7 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.3 | 1.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.3 | 3.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.2 | 2.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 1.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.2 | 0.8 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 47.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 2.7 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.2 | 11.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 0.6 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 1.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 3.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 0.7 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.2 | 5.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 1.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 1.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 3.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.2 | 1.7 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.7 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 1.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 2.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 2.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.6 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 1.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 4.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 22.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 1.8 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.1 | 2.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 3.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 3.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.5 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.1 | 1.2 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 1.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.3 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 4.8 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 0.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 2.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 3.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 2.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 2.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 4.9 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.1 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 5.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 3.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 3.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.1 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 3.3 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 1.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 4.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.6 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 1.5 | GO:0004532 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.5 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 4.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 1.5 | GO:0019956 | chemokine binding(GO:0019956) C-C chemokine binding(GO:0019957) |
| 0.0 | 1.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 3.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 1.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 2.0 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 1.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 9.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 2.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 2.1 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 16.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.4 | 3.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 8.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 2.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 5.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 2.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 15.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 1.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 4.0 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 3.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 0.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 2.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 2.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 12.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.7 | 11.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.6 | 3.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.5 | 4.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.5 | 9.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.4 | 3.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.4 | 16.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 5.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.3 | 4.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.3 | 5.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.3 | 13.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 4.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 3.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.8 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 2.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 9.2 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 0.9 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 1.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 4.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 3.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 1.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 3.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.0 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 2.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |