Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
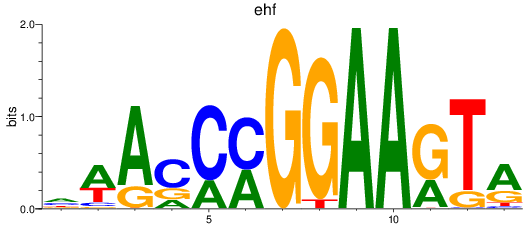
Results for ehf
Z-value: 1.21
Transcription factors associated with ehf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ehf
|
ENSDARG00000052115 | ets homologous factor |
|
ehf
|
ENSDARG00000112589 | ets homologous factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ehf | dr11_v1_chr25_+_10416583_10416583 | 0.46 | 2.8e-06 | Click! |
Activity profile of ehf motif
Sorted Z-values of ehf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_45177373 | 9.05 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr16_-_45001842 | 6.49 |
ENSDART00000037797
|
sult2st3
|
sulfotransferase family 2, cytosolic sulfotransferase 3 |
| chr7_+_45975537 | 6.20 |
ENSDART00000170253
|
plekhf1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
| chr2_+_20539402 | 5.21 |
ENSDART00000129585
|
si:ch73-14h1.2
|
si:ch73-14h1.2 |
| chr5_-_30615901 | 5.17 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr3_+_27713610 | 4.93 |
ENSDART00000019004
|
arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr14_+_28438947 | 4.85 |
ENSDART00000006489
|
acsl4a
|
acyl-CoA synthetase long chain family member 4a |
| chr21_+_25765734 | 4.58 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr20_-_37629084 | 4.43 |
ENSDART00000141734
|
hivep2a
|
human immunodeficiency virus type I enhancer binding protein 2a |
| chr7_-_19923249 | 4.32 |
ENSDART00000078694
|
zgc:110591
|
zgc:110591 |
| chr15_-_2652640 | 4.30 |
ENSDART00000146094
|
cldnf
|
claudin f |
| chr7_+_38762043 | 4.30 |
ENSDART00000036461
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr15_-_29598679 | 4.20 |
ENSDART00000155153
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr25_-_37338048 | 4.09 |
ENSDART00000073439
|
trim44
|
tripartite motif containing 44 |
| chr23_+_20803270 | 4.08 |
ENSDART00000097381
|
zgc:154075
|
zgc:154075 |
| chr17_-_13026634 | 4.03 |
ENSDART00000113713
|
fam177a1
|
family with sequence similarity 177, member A1 |
| chr3_+_36424055 | 4.02 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr18_-_48500545 | 4.00 |
ENSDART00000058989
ENSDART00000098450 |
kcnj1a.6
kcnj1a.5
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 5 |
| chr24_+_35911020 | 3.98 |
ENSDART00000088480
|
abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr1_-_45157243 | 3.95 |
ENSDART00000131882
|
mucms1
|
mucin, multiple PTS and SEA group, member 1 |
| chr20_-_5369105 | 3.84 |
ENSDART00000114316
|
sptlc2b
|
serine palmitoyltransferase, long chain base subunit 2b |
| chr25_+_7784582 | 3.84 |
ENSDART00000155016
|
dgkzb
|
diacylglycerol kinase, zeta b |
| chr6_-_49063085 | 3.84 |
ENSDART00000156124
|
si:ch211-105j21.9
|
si:ch211-105j21.9 |
| chr5_-_34616599 | 3.83 |
ENSDART00000050271
ENSDART00000097975 |
hexb
|
hexosaminidase B (beta polypeptide) |
| chr20_+_22666548 | 3.72 |
ENSDART00000147520
|
lnx1
|
ligand of numb-protein X 1 |
| chr15_-_23793641 | 3.71 |
ENSDART00000122891
|
tmem97
|
transmembrane protein 97 |
| chr1_+_40237276 | 3.70 |
ENSDART00000037553
|
faah2a
|
fatty acid amide hydrolase 2a |
| chr3_-_5067585 | 3.69 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr23_-_31969786 | 3.69 |
ENSDART00000134550
|
ormdl2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr5_+_28271412 | 3.63 |
ENSDART00000031727
|
vamp8
|
vesicle-associated membrane protein 8 (endobrevin) |
| chr1_-_69444 | 3.57 |
ENSDART00000166954
|
si:zfos-1011f11.1
|
si:zfos-1011f11.1 |
| chr19_-_1871415 | 3.51 |
ENSDART00000004585
|
clptm1l
|
CLPTM1-like |
| chr6_-_40768654 | 3.42 |
ENSDART00000184668
ENSDART00000146470 |
arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr2_-_27609189 | 3.39 |
ENSDART00000040555
|
lyn
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr9_+_9502610 | 3.34 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr5_+_63390315 | 3.33 |
ENSDART00000124616
|
rab14
|
RAB14, member RAS oncogene family |
| chr19_-_32500373 | 3.29 |
ENSDART00000052104
|
fuca1.1
|
alpha-L-fucosidase 1, tandem duplicate 1 |
| chr20_-_30900947 | 3.24 |
ENSDART00000153419
ENSDART00000062536 |
hebp2
|
heme binding protein 2 |
| chr22_-_16042243 | 3.21 |
ENSDART00000062633
|
s1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr13_+_27316934 | 3.21 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr15_+_36054864 | 3.20 |
ENSDART00000156697
|
col4a3
|
collagen, type IV, alpha 3 |
| chr25_+_22216723 | 3.20 |
ENSDART00000051291
|
hexa
|
hexosaminidase A (alpha polypeptide) |
| chr8_+_19356072 | 3.19 |
ENSDART00000063272
|
mpeg1.2
|
macrophage expressed 1, tandem duplicate 2 |
| chr13_+_27316632 | 3.17 |
ENSDART00000016121
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr19_-_340347 | 3.15 |
ENSDART00000139924
|
golph3l
|
golgi phosphoprotein 3-like |
| chr3_-_36440705 | 3.15 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr17_+_25331576 | 3.11 |
ENSDART00000157309
|
tmem54a
|
transmembrane protein 54a |
| chr9_-_33081781 | 3.09 |
ENSDART00000165748
|
zgc:172053
|
zgc:172053 |
| chr24_-_6078222 | 3.08 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr4_-_12978925 | 3.07 |
ENSDART00000013839
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr18_+_22994113 | 3.07 |
ENSDART00000171871
|
cbfb
|
core-binding factor, beta subunit |
| chr3_+_34120191 | 3.06 |
ENSDART00000020017
ENSDART00000151700 |
aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr9_-_33081978 | 3.05 |
ENSDART00000100918
|
zgc:172053
|
zgc:172053 |
| chr3_+_24190207 | 3.03 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr3_+_46479705 | 2.98 |
ENSDART00000181564
|
tyk2
|
tyrosine kinase 2 |
| chr15_-_28618502 | 2.94 |
ENSDART00000086902
|
slc6a4a
|
solute carrier family 6 (neurotransmitter transporter), member 4a |
| chr2_+_31838442 | 2.93 |
ENSDART00000066789
|
stard3nl
|
STARD3 N-terminal like |
| chr23_+_20669149 | 2.93 |
ENSDART00000138936
|
arhgef3l
|
Rho guanine nucleotide exchange factor (GEF) 3, like |
| chr1_+_33558555 | 2.92 |
ENSDART00000018472
|
chmp2bb
|
charged multivesicular body protein 2Bb |
| chr5_+_21931124 | 2.92 |
ENSDART00000137627
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr19_-_340641 | 2.90 |
ENSDART00000183848
|
golph3l
|
golgi phosphoprotein 3-like |
| chr10_+_26667475 | 2.90 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr19_+_33093395 | 2.90 |
ENSDART00000019459
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr19_-_20430892 | 2.89 |
ENSDART00000111409
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr3_+_36616713 | 2.87 |
ENSDART00000158284
|
pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr3_+_26223376 | 2.87 |
ENSDART00000128284
|
nudt9
|
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
| chr1_+_51386649 | 2.86 |
ENSDART00000152289
|
atg4da
|
autophagy related 4D, cysteine peptidase a |
| chr24_+_20916751 | 2.84 |
ENSDART00000043193
|
cst14a.1
|
cystatin 14a, tandem duplicate 1 |
| chr5_-_31712399 | 2.82 |
ENSDART00000141328
|
pip5kl1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chr14_+_30272891 | 2.82 |
ENSDART00000017122
|
asah1a
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1a |
| chr22_+_10215558 | 2.81 |
ENSDART00000063274
|
kctd6a
|
potassium channel tetramerization domain containing 6a |
| chr18_+_21071023 | 2.81 |
ENSDART00000100778
|
arpin
|
actin related protein 2/3 complex inhibitor |
| chr16_-_31756859 | 2.78 |
ENSDART00000149170
ENSDART00000126617 ENSDART00000182722 |
ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr17_-_6508406 | 2.77 |
ENSDART00000002778
|
dnajc5gb
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma b |
| chr14_+_31865099 | 2.77 |
ENSDART00000189124
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr24_+_17256793 | 2.76 |
ENSDART00000066764
|
commd3
|
COMM domain containing 3 |
| chr1_-_8020589 | 2.74 |
ENSDART00000143881
|
si:dkeyp-9d4.2
|
si:dkeyp-9d4.2 |
| chr20_-_34127415 | 2.71 |
ENSDART00000010028
|
ptgs2b
|
prostaglandin-endoperoxide synthase 2b |
| chr5_+_23024033 | 2.71 |
ENSDART00000171199
|
chmp1b
|
chromatin modifying protein 1B |
| chr17_-_50311009 | 2.69 |
ENSDART00000153653
|
si:ch73-50f9.4
|
si:ch73-50f9.4 |
| chr14_+_33427837 | 2.68 |
ENSDART00000105687
|
zbtb33
|
zinc finger and BTB domain containing 33 |
| chr5_-_30516646 | 2.67 |
ENSDART00000014666
|
arcn1a
|
archain 1a |
| chr7_-_58178980 | 2.67 |
ENSDART00000073635
|
nsmaf
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr6_+_19689464 | 2.65 |
ENSDART00000164960
|
stx8
|
syntaxin 8 |
| chr17_-_50010121 | 2.64 |
ENSDART00000122747
|
tmem30aa
|
transmembrane protein 30Aa |
| chr19_-_15420678 | 2.62 |
ENSDART00000151454
ENSDART00000027697 |
serinc2
|
serine incorporator 2 |
| chr2_-_47620806 | 2.59 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr20_-_3238110 | 2.58 |
ENSDART00000008077
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr20_+_35282682 | 2.56 |
ENSDART00000187199
|
fam49a
|
family with sequence similarity 49, member A |
| chr11_+_8129536 | 2.56 |
ENSDART00000158112
ENSDART00000011183 |
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr24_+_9590188 | 2.55 |
ENSDART00000137092
|
si:dkey-96n2.1
|
si:dkey-96n2.1 |
| chr19_-_32493866 | 2.55 |
ENSDART00000052090
|
fuca1.2
|
alpha-L-fucosidase 1, tandem duplicate 2 |
| chr16_-_25233515 | 2.55 |
ENSDART00000058943
|
zgc:110182
|
zgc:110182 |
| chr25_+_34749812 | 2.55 |
ENSDART00000185712
|
wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr20_-_14462995 | 2.53 |
ENSDART00000152418
ENSDART00000044125 |
grcc10
|
gene rich cluster, C10 gene |
| chr13_+_24402406 | 2.52 |
ENSDART00000043002
|
rab1ab
|
RAB1A, member RAS oncogene family b |
| chr6_+_49095646 | 2.51 |
ENSDART00000103385
|
slc25a55a
|
solute carrier family 25, member 55a |
| chr14_+_31865324 | 2.51 |
ENSDART00000039880
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr5_+_22459087 | 2.50 |
ENSDART00000134781
|
BX546499.1
|
|
| chr23_+_32021803 | 2.49 |
ENSDART00000012963
|
trappc6b
|
trafficking protein particle complex 6b |
| chr6_+_18251140 | 2.48 |
ENSDART00000169752
|
ccdc40
|
coiled-coil domain containing 40 |
| chr10_+_25369254 | 2.48 |
ENSDART00000164375
|
bach1b
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 b |
| chr4_-_22311610 | 2.47 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr14_+_24277556 | 2.47 |
ENSDART00000122660
|
hnrnpa0a
|
heterogeneous nuclear ribonucleoprotein A0a |
| chr15_+_7064819 | 2.44 |
ENSDART00000155268
|
pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr1_+_12763461 | 2.44 |
ENSDART00000159226
ENSDART00000180121 |
pcdh10a
|
protocadherin 10a |
| chr2_+_43920461 | 2.42 |
ENSDART00000123673
|
si:ch211-195h23.3
|
si:ch211-195h23.3 |
| chr15_-_29598444 | 2.41 |
ENSDART00000154847
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr22_-_8306743 | 2.38 |
ENSDART00000123982
|
CABZ01077218.1
|
|
| chr6_+_25215944 | 2.38 |
ENSDART00000165456
|
si:ch73-97h19.2
|
si:ch73-97h19.2 |
| chr21_+_13908858 | 2.38 |
ENSDART00000148199
|
stxbp1a
|
syntaxin binding protein 1a |
| chr8_-_17987547 | 2.37 |
ENSDART00000112699
ENSDART00000061747 |
fpgt
|
fucose-1-phosphate guanylyltransferase |
| chr24_-_8409641 | 2.34 |
ENSDART00000149662
ENSDART00000149025 |
slc35b3
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B3 |
| chr7_-_48805181 | 2.31 |
ENSDART00000015884
|
mfge8a
|
milk fat globule-EGF factor 8 protein a |
| chr8_-_31107537 | 2.30 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr17_-_20218357 | 2.30 |
ENSDART00000155990
ENSDART00000155632 ENSDART00000156540 ENSDART00000063523 |
mgmt
|
O-6-methylguanine-DNA methyltransferase |
| chr13_-_24311628 | 2.29 |
ENSDART00000004420
|
rab4a
|
RAB4a, member RAS oncogene family |
| chr25_-_25575717 | 2.29 |
ENSDART00000067138
|
hic1l
|
hypermethylated in cancer 1 like |
| chr8_+_52515188 | 2.28 |
ENSDART00000163668
|
si:ch1073-392o20.2
|
si:ch1073-392o20.2 |
| chr1_+_34685405 | 2.28 |
ENSDART00000037986
ENSDART00000166007 |
gpalpp1
|
GPALPP motifs containing 1 |
| chr19_+_40350468 | 2.28 |
ENSDART00000087444
|
hepacam2
|
HEPACAM family member 2 |
| chr16_+_43368572 | 2.26 |
ENSDART00000032778
ENSDART00000193897 |
rnf144b
|
ring finger protein 144B |
| chr5_+_25084385 | 2.26 |
ENSDART00000134526
ENSDART00000111863 |
paxx
|
PAXX, non-homologous end joining factor |
| chr5_-_31470426 | 2.25 |
ENSDART00000002866
ENSDART00000181225 |
p2rx1
|
purinergic receptor P2X, ligand-gated ion channel, 1 |
| chr8_-_39838660 | 2.25 |
ENSDART00000139266
|
ftr98
|
finTRIM family, member 98 |
| chr22_+_5687615 | 2.23 |
ENSDART00000133241
ENSDART00000019854 ENSDART00000138102 |
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr13_+_34689663 | 2.23 |
ENSDART00000133661
|
tasp1
|
taspase, threonine aspartase, 1 |
| chr5_+_19867794 | 2.21 |
ENSDART00000051614
|
tchp
|
trichoplein, keratin filament binding |
| chr21_+_21743599 | 2.20 |
ENSDART00000101700
|
pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr6_-_41085692 | 2.17 |
ENSDART00000181463
|
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr22_+_17399124 | 2.12 |
ENSDART00000145769
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr25_-_35182347 | 2.10 |
ENSDART00000115210
|
ano9a
|
anoctamin 9a |
| chr21_+_5508167 | 2.09 |
ENSDART00000157702
|
ly6m4
|
lymphocyte antigen 6 family member M4 |
| chr2_+_47471943 | 2.09 |
ENSDART00000141974
|
acsl3b
|
acyl-CoA synthetase long chain family member 3b |
| chr13_-_32626247 | 2.06 |
ENSDART00000100663
|
wdr35
|
WD repeat domain 35 |
| chr13_-_22961605 | 2.06 |
ENSDART00000143112
ENSDART00000057641 |
tspan15
|
tetraspanin 15 |
| chr24_-_10897511 | 2.06 |
ENSDART00000145593
ENSDART00000102484 ENSDART00000066784 |
fam49bb
|
family with sequence similarity 49, member Bb |
| chr24_-_24271629 | 2.05 |
ENSDART00000135060
|
rps6ka3b
|
ribosomal protein S6 kinase, polypeptide 3b |
| chr21_-_2173300 | 2.04 |
ENSDART00000172276
|
zgc:163077
|
zgc:163077 |
| chr11_+_11719575 | 2.04 |
ENSDART00000003891
|
jupa
|
junction plakoglobin a |
| chr15_+_41919484 | 2.04 |
ENSDART00000099821
ENSDART00000146246 |
nlrp16
|
NACHT, LRR and PYD domains-containing protein 16 |
| chr19_-_27006764 | 2.02 |
ENSDART00000089540
|
sacm1la
|
SAC1 like phosphatidylinositide phosphatase a |
| chr14_+_13453130 | 2.01 |
ENSDART00000054849
|
pls3
|
plastin 3 (T isoform) |
| chr2_-_43135700 | 2.01 |
ENSDART00000098284
|
ftr14
|
finTRIM family, member 14 |
| chr13_-_25408387 | 2.00 |
ENSDART00000002741
|
itprip
|
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr12_+_31422557 | 2.00 |
ENSDART00000153179
|
zdhhc6
|
zinc finger, DHHC-type containing 6 |
| chr14_-_30897177 | 1.99 |
ENSDART00000087918
|
slc7a3b
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3b |
| chr6_-_21988375 | 1.99 |
ENSDART00000161257
|
plxnb1b
|
plexin b1b |
| chr7_-_8022741 | 1.99 |
ENSDART00000172841
|
si:ch211-163c2.1
|
si:ch211-163c2.1 |
| chr7_+_48805534 | 1.98 |
ENSDART00000145375
ENSDART00000148744 |
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr21_-_35419486 | 1.98 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr2_-_24348948 | 1.98 |
ENSDART00000136559
|
ano8a
|
anoctamin 8a |
| chr4_-_992063 | 1.97 |
ENSDART00000181630
ENSDART00000183898 ENSDART00000160902 |
naga
|
N-acetylgalactosaminidase, alpha |
| chr6_-_40713183 | 1.96 |
ENSDART00000157113
ENSDART00000154810 ENSDART00000153702 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr16_+_38119004 | 1.96 |
ENSDART00000132087
|
pogzb
|
pogo transposable element derived with ZNF domain b |
| chr6_-_41085443 | 1.95 |
ENSDART00000143944
ENSDART00000028217 |
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr21_-_2172924 | 1.95 |
ENSDART00000169262
|
zgc:163077
|
zgc:163077 |
| chr1_+_40034061 | 1.95 |
ENSDART00000011727
|
ccdc149b
|
coiled-coil domain containing 149b |
| chr24_-_24959607 | 1.95 |
ENSDART00000146930
ENSDART00000184482 |
pdk3a
|
pyruvate dehydrogenase kinase, isozyme 3a |
| chr2_+_47471647 | 1.94 |
ENSDART00000184199
|
acsl3b
|
acyl-CoA synthetase long chain family member 3b |
| chr21_-_7035599 | 1.94 |
ENSDART00000139777
|
si:ch211-93g21.1
|
si:ch211-93g21.1 |
| chr13_+_28705143 | 1.94 |
ENSDART00000183338
|
ldb1a
|
LIM domain binding 1a |
| chr5_-_30079434 | 1.94 |
ENSDART00000133981
|
bco2a
|
beta-carotene oxygenase 2a |
| chr18_+_36582815 | 1.93 |
ENSDART00000059311
|
siae
|
sialic acid acetylesterase |
| chr20_+_23947004 | 1.93 |
ENSDART00000144195
|
casp8ap2
|
caspase 8 associated protein 2 |
| chr15_-_41734639 | 1.93 |
ENSDART00000154230
ENSDART00000167443 |
ftr90
|
finTRIM family, member 90 |
| chr8_-_52715911 | 1.93 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr20_-_18382708 | 1.93 |
ENSDART00000170864
ENSDART00000166762 ENSDART00000191333 |
vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr15_+_36445350 | 1.92 |
ENSDART00000154552
|
si:dkey-262k9.2
|
si:dkey-262k9.2 |
| chr1_+_45740183 | 1.91 |
ENSDART00000149116
|
si:ch211-214c7.4
|
si:ch211-214c7.4 |
| chr1_+_55293424 | 1.91 |
ENSDART00000152464
|
zgc:172106
|
zgc:172106 |
| chr3_-_21061931 | 1.91 |
ENSDART00000036741
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr17_-_24890843 | 1.91 |
ENSDART00000184984
ENSDART00000135569 ENSDART00000193661 |
gale
|
UDP-galactose-4-epimerase |
| chr3_+_24681855 | 1.90 |
ENSDART00000170809
|
slc25a17
|
solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein), member 17 |
| chr14_+_20893065 | 1.90 |
ENSDART00000079452
|
lygl1
|
lysozyme g-like 1 |
| chr3_-_40945710 | 1.89 |
ENSDART00000138719
ENSDART00000102416 |
cyp3c4
|
cytochrome P450, family 3, subfamily C, polypeptide 4 |
| chr4_+_9612574 | 1.89 |
ENSDART00000150336
ENSDART00000041289 ENSDART00000150828 |
tmem243b
|
transmembrane protein 243, mitochondrial b |
| chr7_-_58178807 | 1.88 |
ENSDART00000188531
|
nsmaf
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr10_+_36439293 | 1.88 |
ENSDART00000043802
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr10_+_36441124 | 1.87 |
ENSDART00000185626
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr22_-_6777976 | 1.86 |
ENSDART00000187946
|
CT583625.5
|
|
| chr17_-_42842129 | 1.86 |
ENSDART00000009528
|
mocs3
|
molybdenum cofactor synthesis 3 |
| chr7_+_7019911 | 1.86 |
ENSDART00000172421
|
rbm14b
|
RNA binding motif protein 14b |
| chr21_+_45496442 | 1.86 |
ENSDART00000164880
|
si:dkey-223p19.1
|
si:dkey-223p19.1 |
| chr8_-_37249991 | 1.85 |
ENSDART00000189275
ENSDART00000178556 |
rbm39b
|
RNA binding motif protein 39b |
| chr8_+_31248917 | 1.85 |
ENSDART00000112170
|
unm_hu7912
|
un-named hu7912 |
| chr5_+_4366431 | 1.83 |
ENSDART00000168560
ENSDART00000149185 |
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr15_-_28904371 | 1.82 |
ENSDART00000155154
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr4_+_17671614 | 1.81 |
ENSDART00000121714
|
gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr10_+_21789954 | 1.80 |
ENSDART00000157769
ENSDART00000171703 |
pcdh1gc5
|
protocadherin 1 gamma c 5 |
| chr16_+_46401756 | 1.79 |
ENSDART00000147370
ENSDART00000144000 |
rpz2
|
rapunzel 2 |
| chr1_+_36722122 | 1.79 |
ENSDART00000111566
|
tmem184c
|
transmembrane protein 184C |
| chr7_+_26716321 | 1.78 |
ENSDART00000189750
|
cd82a
|
CD82 molecule a |
| chr2_-_56095275 | 1.78 |
ENSDART00000154701
|
si:ch211-178n15.1
|
si:ch211-178n15.1 |
| chr2_-_42415902 | 1.77 |
ENSDART00000142489
|
slco5a1b
|
solute carrier organic anion transporter family member 5A1b |
| chr23_+_31979602 | 1.76 |
ENSDART00000140351
|
pan2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr14_-_30724165 | 1.75 |
ENSDART00000020936
|
fibpa
|
fibroblast growth factor (acidic) intracellular binding protein a |
| chr13_+_11439486 | 1.74 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr5_-_35264517 | 1.74 |
ENSDART00000114981
|
ptcd2
|
pentatricopeptide repeat domain 2 |
| chr24_-_982443 | 1.73 |
ENSDART00000063151
|
napga
|
N-ethylmaleimide-sensitive factor attachment protein, gamma a |
Network of associatons between targets according to the STRING database.
First level regulatory network of ehf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 1.2 | 8.3 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 1.1 | 3.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 1.1 | 6.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.9 | 3.7 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.8 | 3.3 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.8 | 4.9 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.8 | 1.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.7 | 3.7 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.7 | 2.9 | GO:0006837 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.7 | 2.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.7 | 2.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.7 | 2.7 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.7 | 4.0 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.6 | 1.9 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.6 | 3.6 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.5 | 1.6 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.5 | 1.4 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.5 | 1.4 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.5 | 1.4 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.5 | 2.4 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.5 | 1.8 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.5 | 1.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.4 | 1.8 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.4 | 2.0 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.4 | 1.6 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.4 | 1.9 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.4 | 2.6 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.4 | 1.1 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) |
| 0.3 | 3.8 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.3 | 2.1 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.3 | 1.4 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.3 | 3.4 | GO:0009452 | 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.3 | 1.9 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.3 | 1.0 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.3 | 1.6 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.3 | 3.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.3 | 1.9 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.3 | 3.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.3 | 0.9 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.3 | 1.5 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.3 | 1.5 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.3 | 1.5 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.3 | 0.9 | GO:2000639 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.3 | 8.1 | GO:0051923 | sulfation(GO:0051923) |
| 0.3 | 1.2 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.3 | 3.5 | GO:0033198 | response to ATP(GO:0033198) |
| 0.3 | 0.8 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.3 | 3.7 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.3 | 1.4 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
| 0.3 | 1.1 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.3 | 1.4 | GO:0071265 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.3 | 1.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.3 | 1.1 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.3 | 1.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.3 | 1.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 1.3 | GO:0035739 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.3 | 1.0 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.3 | 1.3 | GO:0039528 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.3 | 2.5 | GO:0015800 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.2 | 0.7 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.2 | 2.9 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.2 | 6.8 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.2 | 2.5 | GO:0009791 | post-embryonic development(GO:0009791) |
| 0.2 | 2.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.2 | 1.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.2 | 1.2 | GO:0048714 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.2 | 2.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.2 | 0.6 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.2 | 1.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.2 | 1.7 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 0.6 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.2 | 1.3 | GO:0050820 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.2 | 2.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 | 1.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.2 | 8.9 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.2 | 1.6 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.2 | 0.5 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.2 | 2.4 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.2 | 3.2 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.2 | 1.7 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 1.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.2 | 1.7 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.2 | 1.0 | GO:0089718 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.2 | 1.6 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.2 | 1.6 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.2 | 2.8 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.2 | 2.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 1.7 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 0.9 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.2 | 0.5 | GO:0060945 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.1 | 2.5 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 1.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.9 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.1 | 1.3 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.1 | 1.0 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.8 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 4.0 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.1 | 1.2 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.1 | 1.7 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.1 | 2.2 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 3.1 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.1 | 2.6 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.1 | 2.6 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 2.4 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.5 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 3.0 | GO:0051125 | regulation of actin nucleation(GO:0051125) |
| 0.1 | 1.2 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 1.3 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 1.9 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 0.8 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 1.1 | GO:0061316 | canonical Wnt signaling pathway involved in heart development(GO:0061316) |
| 0.1 | 0.9 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.4 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.1 | 1.5 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 2.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.9 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.9 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 1.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.5 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 2.3 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 2.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.6 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.8 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.1 | 0.8 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 5.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.5 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 1.3 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.1 | 1.1 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.1 | 0.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 1.0 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 3.5 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.1 | 0.6 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 1.3 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 2.1 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 1.3 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 1.2 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.1 | 2.7 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 1.5 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 2.4 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 0.7 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 2.5 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.1 | 0.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 3.2 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.1 | 1.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.5 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 2.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 5.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 5.1 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.1 | 1.0 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 0.7 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 1.2 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 6.5 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.1 | 0.5 | GO:0000479 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.1 | 1.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.2 | GO:0021961 | posterior commissure morphogenesis(GO:0021961) |
| 0.1 | 1.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.4 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 1.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 1.2 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.5 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.0 | 0.5 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 1.6 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 3.3 | GO:0006476 | protein deacetylation(GO:0006476) |
| 0.0 | 1.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 1.1 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 2.3 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 1.0 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 1.1 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.7 | GO:1901655 | cellular response to ketone(GO:1901655) |
| 0.0 | 2.4 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 2.4 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 5.8 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 1.2 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) dendritic spine organization(GO:0097061) |
| 0.0 | 3.0 | GO:0010469 | regulation of receptor activity(GO:0010469) |
| 0.0 | 0.8 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.8 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.0 | 0.6 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 2.0 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 2.4 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 1.3 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 1.1 | GO:1901215 | negative regulation of neuron death(GO:1901215) |
| 0.0 | 0.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 1.7 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.5 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 8.5 | GO:0072657 | protein localization to membrane(GO:0072657) |
| 0.0 | 1.1 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 1.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.8 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 1.3 | GO:0034341 | response to interferon-gamma(GO:0034341) cellular response to interferon-gamma(GO:0071346) |
| 0.0 | 0.9 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) determination of ventral identity(GO:0048264) |
| 0.0 | 0.2 | GO:0034552 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.5 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.8 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.8 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 5.7 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 1.6 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.1 | GO:0003190 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.6 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.8 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.8 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.0 | 1.5 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.1 | GO:0009193 | ribonucleoside diphosphate catabolic process(GO:0009191) pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.0 | 0.4 | GO:0031060 | regulation of histone methylation(GO:0031060) |
| 0.0 | 0.8 | GO:0002218 | activation of innate immune response(GO:0002218) pattern recognition receptor signaling pathway(GO:0002221) innate immune response-activating signal transduction(GO:0002758) |
| 0.0 | 2.6 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 9.2 | GO:0006886 | intracellular protein transport(GO:0006886) |
| 0.0 | 1.0 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.9 | GO:0010970 | establishment of localization by movement along microtubule(GO:0010970) |
| 0.0 | 1.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 5.4 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 1.3 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 1.0 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.4 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 1.4 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 1.8 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 1.2 | GO:0006402 | mRNA catabolic process(GO:0006402) |
| 0.0 | 1.4 | GO:0045216 | cell-cell junction organization(GO:0045216) |
| 0.0 | 0.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.8 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.0 | 0.5 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.4 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.5 | GO:0032868 | response to insulin(GO:0032868) cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 1.1 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.5 | GO:2000146 | negative regulation of cell migration(GO:0030336) negative regulation of cell motility(GO:2000146) |
| 0.0 | 0.7 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 1.0 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.2 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.7 | GO:0016358 | dendrite development(GO:0016358) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.8 | 3.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.6 | 3.8 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.6 | 1.8 | GO:0031251 | PAN complex(GO:0031251) |
| 0.5 | 1.9 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.5 | 1.4 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.5 | 1.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.4 | 2.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.4 | 3.5 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.4 | 3.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.4 | 5.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.4 | 1.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.3 | 1.6 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.3 | 1.9 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.3 | 0.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 0.8 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.2 | 1.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.2 | 1.0 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.2 | 4.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 0.7 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.2 | 1.9 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 0.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 2.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 0.9 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.2 | 3.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 1.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 2.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 2.0 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 0.9 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 1.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 0.5 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.2 | 3.5 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.2 | 1.7 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.2 | 1.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.2 | 2.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 1.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 3.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 2.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 2.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 0.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 1.3 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 2.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.9 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 1.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 2.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 0.6 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 1.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 5.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 6.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.5 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 0.3 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 8.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 2.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 1.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 2.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 1.0 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.1 | 0.8 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 3.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 20.0 | GO:0005764 | lysosome(GO:0005764) |
| 0.1 | 0.6 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 1.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 8.8 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 3.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 3.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 1.9 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 1.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 2.0 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 1.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 6.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 2.4 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 0.6 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.1 | 4.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 2.4 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 2.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 7.8 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.6 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 3.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 4.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 3.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.6 | GO:0030990 | intraciliary transport particle(GO:0030990) intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.9 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 1.3 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 1.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.0 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 7.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 2.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 2.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 12.8 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 1.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0032153 | cell division site(GO:0032153) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 1.5 | 5.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.1 | 4.4 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 1.0 | 2.9 | GO:0015222 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.9 | 2.8 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.8 | 2.4 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.8 | 7.0 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.7 | 2.8 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.7 | 2.7 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.7 | 2.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.7 | 2.0 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.6 | 1.9 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.6 | 1.9 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.6 | 6.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.6 | 2.8 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.5 | 3.8 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.5 | 2.7 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.5 | 4.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.5 | 2.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.5 | 1.6 | GO:0004777 | succinate-semialdehyde dehydrogenase (NAD+) activity(GO:0004777) succinate-semialdehyde dehydrogenase [NAD(P)+] activity(GO:0009013) |
| 0.5 | 2.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.5 | 1.9 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.5 | 1.4 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.5 | 2.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.5 | 1.4 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.4 | 4.0 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.4 | 3.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.4 | 4.9 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.4 | 1.2 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.4 | 1.6 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.3 | 3.5 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.3 | 1.4 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.3 | 1.7 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.3 | 1.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.3 | 1.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.3 | 1.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.3 | 1.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.3 | 3.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.3 | 0.9 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.3 | 1.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.3 | 1.7 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.3 | 3.5 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.3 | 4.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 1.4 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.3 | 1.1 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.3 | 1.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 1.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.3 | 4.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 1.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.3 | 1.8 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.3 | 1.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.3 | 1.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 3.5 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.2 | 1.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 2.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 2.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 2.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.2 | 0.9 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 2.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.2 | 2.9 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.2 | 1.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.2 | 1.5 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.2 | 1.2 | GO:0000995 | transcription factor activity, core RNA polymerase binding(GO:0000990) transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.2 | 2.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 0.7 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.2 | 1.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 1.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 1.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 1.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 1.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 1.5 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.7 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 1.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 1.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 1.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 1.9 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 3.1 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 6.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.7 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 1.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.5 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.1 | 1.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 8.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.4 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 1.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 2.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.3 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 4.1 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 1.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 4.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 2.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.7 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 7.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.4 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 1.0 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 1.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.8 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 2.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.3 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 1.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 1.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.8 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 1.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.5 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 1.3 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 1.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 1.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.2 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 0.9 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 1.4 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.9 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 5.1 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 1.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.9 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 2.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 0.5 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 0.7 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.1 | 2.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 2.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 1.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.7 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 1.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 4.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 3.7 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.9 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 1.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 1.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.5 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.3 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.0 | 0.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 3.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 4.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.7 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 7.0 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 14.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.5 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 1.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 2.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.7 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 1.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.9 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 0.2 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.6 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 2.3 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.2 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 2.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.6 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 2.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.3 | 2.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 3.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 3.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 1.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 2.8 | PID EPO PATHWAY | EPO signaling pathway |
| 0.2 | 3.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.2 | 4.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 4.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.2 | 5.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 4.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 3.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 2.3 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 4.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 0.4 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 2.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.7 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 1.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 2.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 0.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 3.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 0.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 0.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 0.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 1.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 7.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.7 | 6.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.6 | 4.8 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.4 | 3.5 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.3 | 4.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.2 | 3.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 1.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.2 | 1.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 2.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 1.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 2.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 3.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 2.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 1.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 2.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.6 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 1.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.9 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 1.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.9 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 0.6 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 0.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 0.3 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.1 | 4.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 1.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 0.4 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.1 | 1.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 6.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 2.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.0 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.7 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |