Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
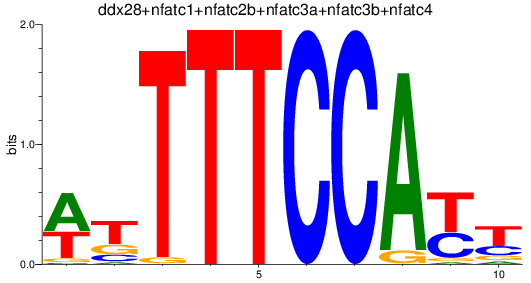
Results for ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4
Z-value: 1.50
Transcription factors associated with ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfatc1
|
ENSDARG00000036168 | nuclear factor of activated T cells 1 |
|
nfatc3b
|
ENSDARG00000051729 | nuclear factor of activated T cells 3b |
|
nfatc4
|
ENSDARG00000054162 | nuclear factor of activated T cells 4 |
|
nfatc3a
|
ENSDARG00000076297 | nuclear factor of activated T cells 3a |
|
nfatc2b
|
ENSDARG00000079972 | nuclear factor of activated T cells 2b |
|
ddx28
|
ENSDARG00000112133 | nuclear factor of activated T cells 3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfatc1 | dr11_v1_chr19_+_22216778_22216778 | 0.61 | 8.1e-11 | Click! |
| nfatc3b | dr11_v1_chr25_-_36248053_36248053 | 0.29 | 4.4e-03 | Click! |
| nfatc2b | dr11_v1_chr6_-_55254786_55254786 | -0.29 | 4.7e-03 | Click! |
| nfatc4 | dr11_v1_chr2_-_37797577_37797577 | 0.12 | 2.3e-01 | Click! |
| nfatc3a | dr11_v1_chr7_-_34927961_34927961 | -0.10 | 3.6e-01 | Click! |
Activity profile of ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4 motif
Sorted Z-values of ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_26064480 | 24.00 |
ENSDART00000158215
ENSDART00000171206 ENSDART00000171212 ENSDART00000182956 ENSDART00000186779 |
ldb3b
|
LIM domain binding 3b |
| chr2_-_24289641 | 15.79 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr8_-_40464935 | 13.72 |
ENSDART00000040013
|
myl7
|
myosin, light chain 7, regulatory |
| chr18_+_26743749 | 12.19 |
ENSDART00000145212
|
alpk3a
|
alpha-kinase 3a |
| chr15_-_20933574 | 12.14 |
ENSDART00000152648
ENSDART00000152448 ENSDART00000152244 |
usp2a
|
ubiquitin specific peptidase 2a |
| chr6_-_39313027 | 12.03 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr13_+_23157053 | 11.80 |
ENSDART00000162359
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr2_+_42191592 | 11.52 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr3_-_32817274 | 11.08 |
ENSDART00000142582
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr9_-_42989297 | 9.97 |
ENSDART00000126871
|
ttn.2
|
titin, tandem duplicate 2 |
| chr9_+_6587056 | 9.55 |
ENSDART00000193421
|
fhl2a
|
four and a half LIM domains 2a |
| chr16_-_22713152 | 9.49 |
ENSDART00000140953
ENSDART00000143836 |
si:ch211-105c13.3
|
si:ch211-105c13.3 |
| chr3_-_61181018 | 9.33 |
ENSDART00000187970
|
pvalb4
|
parvalbumin 4 |
| chr3_-_39152478 | 9.22 |
ENSDART00000154550
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr19_+_5674907 | 9.05 |
ENSDART00000042189
|
pdk2b
|
pyruvate dehydrogenase kinase, isozyme 2b |
| chr9_+_6587364 | 9.04 |
ENSDART00000122279
|
fhl2a
|
four and a half LIM domains 2a |
| chr1_-_38195012 | 8.83 |
ENSDART00000020409
|
hand2
|
heart and neural crest derivatives expressed 2 |
| chr6_-_10233538 | 8.78 |
ENSDART00000182004
ENSDART00000149237 ENSDART00000148876 |
xirp2a
|
xin actin binding repeat containing 2a |
| chr18_+_26719787 | 8.77 |
ENSDART00000141672
|
alpk3a
|
alpha-kinase 3a |
| chr16_+_23397785 | 8.76 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr14_+_49296052 | 8.73 |
ENSDART00000006073
ENSDART00000105346 |
anxa6
|
annexin A6 |
| chr11_-_29946640 | 8.67 |
ENSDART00000079175
|
zgc:113276
|
zgc:113276 |
| chr3_-_20091964 | 8.56 |
ENSDART00000029386
ENSDART00000020253 ENSDART00000124326 |
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr25_-_28674739 | 8.44 |
ENSDART00000067073
|
lrrc10
|
leucine rich repeat containing 10 |
| chr7_+_65564163 | 8.37 |
ENSDART00000082679
|
mical2a
|
microtubule associated monooxygenase, calponin and LIM domain containing 2a |
| chr22_-_15587360 | 8.23 |
ENSDART00000142717
ENSDART00000138978 |
tpm4a
|
tropomyosin 4a |
| chr1_-_38816685 | 8.17 |
ENSDART00000075230
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr7_-_38658411 | 8.08 |
ENSDART00000109463
ENSDART00000017155 |
npsn
|
nephrosin |
| chr16_-_12723324 | 7.98 |
ENSDART00000131915
|
sbk3
|
SH3 domain binding kinase family, member 3 |
| chr16_+_23398369 | 7.95 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr25_+_14087045 | 7.79 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr24_+_35975398 | 7.62 |
ENSDART00000173058
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr11_-_11471857 | 7.59 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr3_-_19367081 | 7.58 |
ENSDART00000191369
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr1_-_41192059 | 7.58 |
ENSDART00000084665
ENSDART00000135369 |
dok7
|
docking protein 7 |
| chr2_-_20923864 | 6.83 |
ENSDART00000006870
|
ptgs2a
|
prostaglandin-endoperoxide synthase 2a |
| chr11_+_13630107 | 6.81 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr11_-_30611814 | 6.67 |
ENSDART00000089803
|
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr7_-_21905851 | 6.57 |
ENSDART00000111066
ENSDART00000020288 |
epoa
|
erythropoietin a |
| chr23_+_13814978 | 6.41 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr6_+_60112200 | 6.31 |
ENSDART00000008243
|
prelid3b
|
PRELI domain containing 3 |
| chr6_-_48094342 | 6.30 |
ENSDART00000137458
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr22_-_26353916 | 6.20 |
ENSDART00000077958
|
capn2b
|
calpain 2, (m/II) large subunit b |
| chr25_-_32888115 | 6.19 |
ENSDART00000087586
|
c2cd4a
|
C2 calcium dependent domain containing 4A |
| chr9_+_35860975 | 6.06 |
ENSDART00000134447
|
rcan1a
|
regulator of calcineurin 1a |
| chr22_+_28446557 | 5.98 |
ENSDART00000089546
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr22_+_28446365 | 5.92 |
ENSDART00000189359
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr11_-_30612166 | 5.90 |
ENSDART00000177984
|
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr21_-_22928214 | 5.83 |
ENSDART00000182760
|
dub
|
duboraya |
| chr20_+_28364742 | 5.77 |
ENSDART00000103355
|
rhov
|
ras homolog family member V |
| chr3_+_14463941 | 5.75 |
ENSDART00000170927
|
cnn1b
|
calponin 1, basic, smooth muscle, b |
| chr13_+_13681681 | 5.70 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr16_-_35975254 | 5.66 |
ENSDART00000167537
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr9_-_23990416 | 5.58 |
ENSDART00000113176
|
col6a3
|
collagen, type VI, alpha 3 |
| chr21_+_21621042 | 5.56 |
ENSDART00000134907
|
tgfb1b
|
transforming growth factor, beta 1b |
| chr16_+_25245857 | 5.50 |
ENSDART00000155220
|
klhl38b
|
kelch-like family member 38b |
| chr10_-_43771447 | 5.27 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr11_+_10541258 | 5.26 |
ENSDART00000132365
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr9_+_33145522 | 5.23 |
ENSDART00000005879
|
atp5po
|
ATP synthase peripheral stalk subunit OSCP |
| chr18_+_26750516 | 5.22 |
ENSDART00000110843
|
alpk3a
|
alpha-kinase 3a |
| chr17_+_24848976 | 5.19 |
ENSDART00000062917
|
cx35.4
|
connexin 35.4 |
| chr14_+_26759332 | 5.00 |
ENSDART00000088484
|
ahnak
|
AHNAK nucleoprotein |
| chr7_-_52558495 | 4.98 |
ENSDART00000138263
ENSDART00000009938 ENSDART00000174292 ENSDART00000174218 ENSDART00000174335 |
tcf12
|
transcription factor 12 |
| chr1_-_58664854 | 4.94 |
ENSDART00000109528
|
adgre5b.3
|
adhesion G protein-coupled receptor E5b, duplicate 3 |
| chr7_-_52417777 | 4.94 |
ENSDART00000110265
|
myzap
|
myocardial zonula adherens protein |
| chr11_-_30601401 | 4.93 |
ENSDART00000172244
ENSDART00000046800 |
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr4_+_7677318 | 4.92 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr6_+_49095646 | 4.89 |
ENSDART00000103385
|
slc25a55a
|
solute carrier family 25, member 55a |
| chr20_+_29436601 | 4.83 |
ENSDART00000136804
|
fmn1
|
formin 1 |
| chr13_-_13754091 | 4.80 |
ENSDART00000131255
|
ky
|
kyphoscoliosis peptidase |
| chr2_-_24369087 | 4.79 |
ENSDART00000081237
|
plvapa
|
plasmalemma vesicle associated protein a |
| chr12_+_17106117 | 4.68 |
ENSDART00000149990
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr20_-_49681850 | 4.66 |
ENSDART00000025926
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr9_-_23994225 | 4.66 |
ENSDART00000140346
|
col6a3
|
collagen, type VI, alpha 3 |
| chr8_+_27743550 | 4.64 |
ENSDART00000046004
|
wnt2bb
|
wingless-type MMTV integration site family, member 2Bb |
| chr19_+_18797623 | 4.50 |
ENSDART00000166172
|
ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr20_+_53577502 | 4.47 |
ENSDART00000126983
|
myh6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr20_+_46040666 | 4.47 |
ENSDART00000060744
|
si:dkey-7c18.24
|
si:dkey-7c18.24 |
| chr2_+_4207209 | 4.46 |
ENSDART00000157903
ENSDART00000166476 |
gata6
|
GATA binding protein 6 |
| chr2_+_38161318 | 4.43 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr5_+_30179010 | 4.31 |
ENSDART00000134624
|
adamts15a
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15a |
| chr13_-_42749916 | 4.29 |
ENSDART00000140019
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr6_-_34220641 | 4.26 |
ENSDART00000102391
|
dmrt2b
|
doublesex and mab-3 related transcription factor 2b |
| chr25_+_6122823 | 4.24 |
ENSDART00000191824
ENSDART00000067514 |
rbpms2a
|
RNA binding protein with multiple splicing 2a |
| chr16_+_30438041 | 4.22 |
ENSDART00000137977
|
pear1
|
platelet endothelial aggregation receptor 1 |
| chr16_+_35887868 | 4.19 |
ENSDART00000169677
ENSDART00000170772 |
thrap3a
|
thyroid hormone receptor associated protein 3a |
| chr24_+_11334733 | 4.16 |
ENSDART00000147552
ENSDART00000143171 |
si:dkey-12l12.1
|
si:dkey-12l12.1 |
| chr21_+_2091895 | 4.13 |
ENSDART00000161828
|
si:rp71-1h20.9
|
si:rp71-1h20.9 |
| chr21_-_17603182 | 4.04 |
ENSDART00000020048
ENSDART00000177270 |
gsna
|
gelsolin a |
| chr19_-_38611814 | 4.01 |
ENSDART00000151958
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr9_+_31752628 | 3.96 |
ENSDART00000060054
|
itgbl1
|
integrin, beta-like 1 |
| chr23_-_19682971 | 3.88 |
ENSDART00000048891
|
zgc:193598
|
zgc:193598 |
| chr7_-_30367650 | 3.87 |
ENSDART00000075519
|
aldh1a2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr7_+_26167420 | 3.82 |
ENSDART00000173941
|
si:ch211-196f2.6
|
si:ch211-196f2.6 |
| chr22_+_19311411 | 3.81 |
ENSDART00000133234
ENSDART00000138284 |
si:dkey-21e2.16
|
si:dkey-21e2.16 |
| chr5_-_34997630 | 3.80 |
ENSDART00000170684
|
btf3
|
basic transcription factor 3 |
| chr13_-_22646134 | 3.80 |
ENSDART00000136808
|
mmrn2a
|
multimerin 2a |
| chr6_-_42983843 | 3.79 |
ENSDART00000130666
|
tnfrsf18
|
tumor necrosis factor receptor superfamily, member 18 |
| chr6_-_49063085 | 3.75 |
ENSDART00000156124
|
si:ch211-105j21.9
|
si:ch211-105j21.9 |
| chr20_+_6773790 | 3.75 |
ENSDART00000169966
|
igfbp3
|
insulin-like growth factor binding protein 3 |
| chr6_-_33875919 | 3.73 |
ENSDART00000190411
|
tmem69
|
transmembrane protein 69 |
| chr13_+_16279890 | 3.70 |
ENSDART00000101775
ENSDART00000057948 |
anxa11a
|
annexin A11a |
| chr5_+_37091626 | 3.65 |
ENSDART00000161054
|
tagln2
|
transgelin 2 |
| chr6_-_49673476 | 3.54 |
ENSDART00000112226
|
apcdd1l
|
adenomatosis polyposis coli down-regulated 1-like |
| chr21_+_29077509 | 3.52 |
ENSDART00000128561
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr16_+_31542645 | 3.52 |
ENSDART00000163724
|
SLA (1 of many)
|
Src like adaptor |
| chr7_-_52531252 | 3.49 |
ENSDART00000174369
|
tcf12
|
transcription factor 12 |
| chr1_-_29658721 | 3.44 |
ENSDART00000132063
|
si:dkey-1h24.6
|
si:dkey-1h24.6 |
| chr13_+_25410594 | 3.43 |
ENSDART00000112092
|
calhm3
|
calcium homeostasis modulator 3 |
| chr20_+_31287356 | 3.40 |
ENSDART00000007688
|
slc22a16
|
solute carrier family 22 (organic cation/carnitine transporter), member 16 |
| chr20_+_38276690 | 3.38 |
ENSDART00000061437
|
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr6_-_36552844 | 3.34 |
ENSDART00000023613
|
her6
|
hairy-related 6 |
| chr10_+_26612321 | 3.32 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr11_-_31039533 | 3.32 |
ENSDART00000127355
|
ier2b
|
immediate early response 2b |
| chr15_-_34468599 | 3.29 |
ENSDART00000192984
|
meox2a
|
mesenchyme homeobox 2a |
| chr25_-_21822426 | 3.25 |
ENSDART00000151993
|
zgc:158222
|
zgc:158222 |
| chr19_-_33087246 | 3.23 |
ENSDART00000052078
|
klhl38a
|
kelch-like family member 38a |
| chr2_-_30182353 | 3.22 |
ENSDART00000019149
|
rpl7
|
ribosomal protein L7 |
| chr20_+_38285671 | 3.19 |
ENSDART00000061432
|
ccl38a.4
|
chemokine (C-C motif) ligand 38, duplicate 4 |
| chr23_+_36101185 | 3.17 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr8_+_25616946 | 3.12 |
ENSDART00000133983
|
slc38a5a
|
solute carrier family 38, member 5a |
| chr6_-_40899618 | 3.06 |
ENSDART00000153949
ENSDART00000021969 |
zgc:172271
|
zgc:172271 |
| chr20_+_6756247 | 3.04 |
ENSDART00000167344
|
igfbp3
|
insulin-like growth factor binding protein 3 |
| chr5_-_15851953 | 3.00 |
ENSDART00000173101
|
si:dkey-1k23.3
|
si:dkey-1k23.3 |
| chr1_-_54425791 | 2.97 |
ENSDART00000039911
|
pkd1a
|
polycystic kidney disease 1a |
| chr11_-_18705303 | 2.96 |
ENSDART00000059732
|
id1
|
inhibitor of DNA binding 1 |
| chr11_-_40165683 | 2.94 |
ENSDART00000186510
|
si:dkey-264d12.4
|
si:dkey-264d12.4 |
| chr20_+_1996202 | 2.91 |
ENSDART00000184143
|
CABZ01092781.1
|
|
| chr2_-_25140022 | 2.87 |
ENSDART00000134543
|
nceh1a
|
neutral cholesterol ester hydrolase 1a |
| chr22_-_28653074 | 2.86 |
ENSDART00000154717
|
col8a1b
|
collagen, type VIII, alpha 1b |
| chr18_+_13309862 | 2.83 |
ENSDART00000131555
|
si:ch211-260p9.3
|
si:ch211-260p9.3 |
| chr13_-_18835254 | 2.81 |
ENSDART00000147579
ENSDART00000146795 |
lzts2a
|
leucine zipper, putative tumor suppressor 2a |
| chr6_-_52675630 | 2.81 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr1_+_51475094 | 2.79 |
ENSDART00000146352
|
meis1a
|
Meis homeobox 1 a |
| chr15_-_17618800 | 2.78 |
ENSDART00000157185
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr2_+_10914023 | 2.78 |
ENSDART00000146546
|
mrpl37
|
mitochondrial ribosomal protein L37 |
| chr15_-_25518084 | 2.77 |
ENSDART00000158594
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr8_-_45834825 | 2.77 |
ENSDART00000132965
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr23_+_44049509 | 2.77 |
ENSDART00000102003
|
txk
|
TXK tyrosine kinase |
| chr11_-_34628789 | 2.77 |
ENSDART00000192433
|
mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr7_+_44445595 | 2.76 |
ENSDART00000108766
|
cdh5
|
cadherin 5 |
| chr14_+_46028003 | 2.76 |
ENSDART00000113469
|
nocta
|
nocturnin a |
| chr6_-_50204262 | 2.75 |
ENSDART00000163648
|
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr16_+_40301056 | 2.74 |
ENSDART00000058578
|
rspo3
|
R-spondin 3 |
| chr13_-_39947335 | 2.72 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr2_-_48196092 | 2.72 |
ENSDART00000139944
|
snorc
|
secondary ossification center associated regulator of chondrocyte maturation |
| chr14_-_4120636 | 2.68 |
ENSDART00000059230
|
irf2
|
interferon regulatory factor 2 |
| chr11_-_10643091 | 2.66 |
ENSDART00000192314
|
mcf2l2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr16_+_20910186 | 2.66 |
ENSDART00000046766
|
hoxa10b
|
homeobox A10b |
| chr8_+_17078692 | 2.65 |
ENSDART00000023206
|
plk2b
|
polo-like kinase 2b (Drosophila) |
| chr3_-_10677890 | 2.63 |
ENSDART00000155382
ENSDART00000171319 |
si:ch1073-144j5.2
|
si:ch1073-144j5.2 |
| chr12_+_16281312 | 2.63 |
ENSDART00000152500
|
ppp1r3cb
|
protein phosphatase 1, regulatory subunit 3Cb |
| chr14_-_33454595 | 2.61 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr5_-_44496553 | 2.59 |
ENSDART00000178081
|
gas1a
|
growth arrest-specific 1a |
| chr4_-_13502549 | 2.56 |
ENSDART00000140366
|
si:ch211-266a5.12
|
si:ch211-266a5.12 |
| chr14_+_30568961 | 2.56 |
ENSDART00000184303
|
mrpl11
|
mitochondrial ribosomal protein L11 |
| chr7_+_30051880 | 2.54 |
ENSDART00000075609
|
pstpip1b
|
proline-serine-threonine phosphatase interacting protein 1b |
| chr17_+_27456804 | 2.54 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr10_-_7892192 | 2.53 |
ENSDART00000145480
|
smtna
|
smoothelin a |
| chr13_+_35690023 | 2.53 |
ENSDART00000128865
ENSDART00000130050 |
psme4a
|
proteasome activator subunit 4a |
| chr10_+_42159471 | 2.53 |
ENSDART00000122160
|
CR925773.1
|
|
| chr8_-_38403018 | 2.52 |
ENSDART00000134100
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr21_-_43131752 | 2.52 |
ENSDART00000024137
|
p4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide 2 |
| chr23_-_764135 | 2.51 |
ENSDART00000010248
ENSDART00000183978 |
mitfb
|
microphthalmia-associated transcription factor b |
| chr22_-_10470663 | 2.51 |
ENSDART00000143352
|
omd
|
osteomodulin |
| chr20_-_13660600 | 2.50 |
ENSDART00000063826
|
TAGAP
|
si:ch211-122h15.4 |
| chr17_-_25382367 | 2.49 |
ENSDART00000162306
ENSDART00000165282 |
lck
|
LCK proto-oncogene, Src family tyrosine kinase |
| chr8_+_40628926 | 2.48 |
ENSDART00000163598
|
dusp2
|
dual specificity phosphatase 2 |
| chr8_-_50132860 | 2.46 |
ENSDART00000149964
|
antxr1a
|
anthrax toxin receptor 1a |
| chr10_+_8437930 | 2.45 |
ENSDART00000074553
|
pptc7b
|
PTC7 protein phosphatase homolog b |
| chr14_+_6182346 | 2.44 |
ENSDART00000075525
|
si:ch73-22a13.3
|
si:ch73-22a13.3 |
| chr25_-_12635371 | 2.43 |
ENSDART00000162463
|
zcchc14
|
zinc finger, CCHC domain containing 14 |
| chr9_+_13985567 | 2.43 |
ENSDART00000102296
|
cd28
|
CD28 molecule |
| chr20_+_38279523 | 2.38 |
ENSDART00000061311
|
ccl38a.5
|
chemokine (C-C motif) ligand 38, duplicate 5 |
| chr8_-_19342111 | 2.35 |
ENSDART00000138881
|
rgl1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr6_+_48978202 | 2.30 |
ENSDART00000150023
|
slc16a1a
|
solute carrier family 16 (monocarboxylate transporter), member 1a |
| chr19_-_35596207 | 2.30 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr6_+_52263236 | 2.29 |
ENSDART00000144174
|
esyt1b
|
extended synaptotagmin-like protein 1b |
| chr12_+_20627505 | 2.26 |
ENSDART00000074384
|
stx4
|
syntaxin 4 |
| chr16_-_28091597 | 2.26 |
ENSDART00000166595
|
CR626941.1
|
|
| chr1_-_44484 | 2.26 |
ENSDART00000171547
ENSDART00000164075 ENSDART00000168091 |
tmem39a
|
transmembrane protein 39A |
| chr10_+_8767541 | 2.24 |
ENSDART00000170272
|
itga1
|
integrin, alpha 1 |
| chr3_-_34076807 | 2.21 |
ENSDART00000150964
|
ighv7-1
|
immunoglobulin heavy variable 7-1 |
| chr20_-_13660767 | 2.20 |
ENSDART00000127654
|
TAGAP
|
si:ch211-122h15.4 |
| chr15_-_21852963 | 2.18 |
ENSDART00000153581
ENSDART00000155588 ENSDART00000079399 |
laynb
|
layilin b |
| chr22_+_18884971 | 2.18 |
ENSDART00000186332
|
fstl3
|
follistatin-like 3 (secreted glycoprotein) |
| chr16_-_17541890 | 2.18 |
ENSDART00000131328
|
clcn1b
|
chloride channel, voltage-sensitive 1b |
| chr5_+_33498253 | 2.17 |
ENSDART00000140993
|
ms4a17c.2
|
membrane-spanning 4-domains, subfamily A, member 17c.2 |
| chr22_+_5752257 | 2.17 |
ENSDART00000143052
|
cox14
|
COX14 cytochrome c oxidase assembly factor |
| chr9_-_8454060 | 2.14 |
ENSDART00000110158
|
irs2b
|
insulin receptor substrate 2b |
| chr23_-_14825450 | 2.13 |
ENSDART00000188009
|
sla2
|
Src-like-adaptor 2 |
| chr8_-_32803227 | 2.12 |
ENSDART00000110079
|
zgc:194839
|
zgc:194839 |
| chr21_-_27213166 | 2.12 |
ENSDART00000146959
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr3_-_11322732 | 2.11 |
ENSDART00000161957
|
AL935044.2
|
|
| chr25_+_6451038 | 2.10 |
ENSDART00000009971
|
snx33
|
sorting nexin 33 |
| chr4_-_9722568 | 2.09 |
ENSDART00000067190
|
tspan9b
|
tetraspanin 9b |
| chr8_-_19904124 | 2.08 |
ENSDART00000129193
|
trabd2b
|
TraB domain containing 2B |
| chr21_+_30746348 | 2.06 |
ENSDART00000050172
|
trpc2b
|
transient receptor potential cation channel, subfamily C, member 2b |
| chr4_+_76764315 | 2.05 |
ENSDART00000075598
|
ms4a17a.12
|
membrane-spanning 4-domains, subfamily A, member 17A.12 |
| chr9_+_13682133 | 2.03 |
ENSDART00000175639
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
Network of associatons between targets according to the STRING database.
First level regulatory network of ddx28+nfatc1+nfatc2b+nfatc3a+nfatc3b+nfatc4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 18.2 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 2.2 | 8.7 | GO:0001778 | plasma membrane repair(GO:0001778) chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 1.9 | 5.8 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 1.9 | 5.8 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 1.7 | 6.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.6 | 4.7 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 1.4 | 8.4 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 1.4 | 2.7 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 1.3 | 5.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 1.3 | 3.8 | GO:0002727 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 1.2 | 7.2 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 1.2 | 4.6 | GO:0061010 | gall bladder development(GO:0061010) |
| 1.1 | 5.7 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 1.1 | 3.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 1.1 | 4.5 | GO:0045429 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 1.0 | 3.9 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 1.0 | 19.2 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.9 | 6.6 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.9 | 10.0 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.9 | 4.5 | GO:0043011 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.8 | 3.1 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.7 | 3.0 | GO:0032965 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.7 | 6.4 | GO:0051694 | skeletal muscle thin filament assembly(GO:0030240) pointed-end actin filament capping(GO:0051694) |
| 0.7 | 2.8 | GO:0036445 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.7 | 2.8 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.7 | 2.7 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.6 | 8.4 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.6 | 2.5 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.6 | 6.8 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.6 | 1.8 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.6 | 1.7 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.6 | 3.3 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.5 | 4.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.5 | 4.9 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.5 | 1.4 | GO:1900158 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.5 | 2.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.5 | 0.9 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.4 | 1.7 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.4 | 2.5 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.4 | 1.7 | GO:0015871 | choline transport(GO:0015871) |
| 0.4 | 9.5 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.4 | 25.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.4 | 1.6 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.4 | 5.6 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.4 | 8.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.4 | 3.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.4 | 2.8 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 1.7 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.3 | 1.6 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.3 | 2.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.3 | 3.5 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.3 | 1.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.3 | 5.4 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.3 | 2.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.3 | 1.1 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.3 | 1.3 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.2 | 5.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 10.1 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.2 | 12.8 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) |
| 0.2 | 4.7 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.2 | 1.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.2 | 1.0 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.2 | 2.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 1.5 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.2 | 2.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 8.0 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.2 | 2.8 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.2 | 8.5 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.2 | 3.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.2 | 7.6 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.2 | 0.8 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.2 | 0.6 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 4.5 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 1.0 | GO:0050820 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 4.0 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 5.7 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 3.7 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.1 | 2.4 | GO:0070672 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 11.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 4.2 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 5.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 0.5 | GO:0071962 | establishment of mitotic sister chromatid cohesion(GO:0034087) mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.1 | 0.6 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.1 | 1.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.4 | GO:0002312 | B cell activation involved in immune response(GO:0002312) isotype switching(GO:0045190) |
| 0.1 | 0.9 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 3.2 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 1.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.6 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.1 | 1.6 | GO:0045116 | protein neddylation(GO:0045116) positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 3.5 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 1.1 | GO:0030431 | sleep(GO:0030431) |
| 0.1 | 8.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 1.8 | GO:0050926 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.1 | 2.8 | GO:0010634 | positive regulation of epithelial cell migration(GO:0010634) |
| 0.1 | 1.0 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 2.5 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.1 | 2.2 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.1 | 1.0 | GO:0048512 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.1 | 9.1 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 6.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 1.8 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 8.2 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 0.6 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.1 | 0.4 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.1 | 0.8 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.1 | 3.8 | GO:0045785 | positive regulation of cell adhesion(GO:0045785) |
| 0.1 | 1.1 | GO:0030816 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.1 | 2.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 1.9 | GO:0001736 | establishment of planar polarity(GO:0001736) establishment of tissue polarity(GO:0007164) |
| 0.1 | 0.3 | GO:0060350 | endochondral bone morphogenesis(GO:0060350) cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.1 | 8.9 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.1 | 0.5 | GO:0002363 | CD4-positive, alpha-beta T cell differentiation involved in immune response(GO:0002294) T-helper cell lineage commitment(GO:0002295) T cell lineage commitment(GO:0002360) alpha-beta T cell lineage commitment(GO:0002363) acute-phase response(GO:0006953) T-helper cell differentiation(GO:0042093) positive T cell selection(GO:0043368) CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 2.1 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 4.7 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 1.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 2.8 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.1 | 1.5 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 2.6 | GO:0050863 | regulation of T cell activation(GO:0050863) regulation of leukocyte cell-cell adhesion(GO:1903037) |
| 0.1 | 1.4 | GO:0071594 | B cell differentiation(GO:0030183) T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 | 1.6 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.1 | 2.1 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 4.1 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 4.3 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 1.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 2.7 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 2.3 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 2.3 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.1 | 8.1 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 17.2 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 2.8 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.1 | 4.8 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.1 | 30.6 | GO:0030036 | actin cytoskeleton organization(GO:0030036) |
| 0.1 | 0.4 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.1 | 1.6 | GO:0060914 | heart formation(GO:0060914) |
| 0.1 | 1.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.2 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.0 | 1.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 1.0 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 1.5 | GO:2001056 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043280) positive regulation of cysteine-type endopeptidase activity(GO:2001056) |
| 0.0 | 3.8 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.0 | 1.2 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 3.7 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.5 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.3 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 1.5 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 2.8 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 1.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 1.0 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.3 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 2.1 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.6 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 3.4 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.7 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 2.8 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.5 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 4.7 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.5 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.3 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.8 | GO:0050432 | catecholamine secretion(GO:0050432) regulation of catecholamine secretion(GO:0050433) |
| 0.0 | 0.8 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 2.5 | GO:0051216 | cartilage development(GO:0051216) |
| 0.0 | 0.0 | GO:0010893 | positive regulation of steroid biosynthetic process(GO:0010893) positive regulation of steroid metabolic process(GO:0045940) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.2 | GO:0043065 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 0.9 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.2 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) |
| 0.0 | 0.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.9 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 12.0 | GO:0045095 | keratin filament(GO:0045095) |
| 1.0 | 5.0 | GO:0043034 | costamere(GO:0043034) |
| 0.7 | 5.2 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.7 | 14.0 | GO:0031430 | M band(GO:0031430) |
| 0.6 | 3.8 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.6 | 2.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.6 | 26.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.4 | 2.0 | GO:0034657 | GID complex(GO:0034657) |
| 0.3 | 1.7 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.3 | 4.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 11.5 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.3 | 10.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.3 | 2.8 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.3 | 18.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 1.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 18.6 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.2 | 14.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 24.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 14.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 2.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 6.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 6.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 10.0 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 8.5 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 7.9 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 2.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.5 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 9.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 0.5 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 13.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 0.9 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 4.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 20.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.6 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 0.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 3.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.8 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 7.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 49.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.4 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 1.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.8 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 2.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 2.8 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.7 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 4.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.6 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 1.8 | 5.3 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 1.7 | 6.8 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 1.4 | 8.4 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 1.3 | 17.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 1.2 | 24.0 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 1.1 | 9.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.0 | 6.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.9 | 7.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.9 | 26.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.9 | 5.6 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.9 | 6.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.9 | 6.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.8 | 3.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.6 | 6.8 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.6 | 1.7 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.5 | 1.6 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.5 | 4.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.5 | 5.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.4 | 2.6 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.4 | 1.7 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.4 | 4.9 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.4 | 11.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.4 | 8.6 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.4 | 1.6 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 8.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.4 | 3.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.4 | 1.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.4 | 1.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.3 | 2.8 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.3 | 10.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 6.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 3.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.3 | 3.6 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.3 | 3.9 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.3 | 2.2 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.3 | 5.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.3 | 5.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.3 | 1.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.3 | 2.5 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.2 | 0.7 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.2 | 4.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.2 | 0.9 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 2.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.2 | 9.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 2.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 2.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 2.8 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.2 | 1.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 8.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.2 | 2.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 2.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 1.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 6.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 21.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 2.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.6 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 4.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 1.0 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 2.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 2.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.5 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.6 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.1 | 1.0 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.9 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 1.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 34.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.9 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 1.7 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 1.5 | GO:0048018 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.1 | 1.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 7.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 3.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 2.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.8 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 8.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 9.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 4.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 2.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 1.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.6 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 1.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 19.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 16.7 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.1 | 2.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.3 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 9.6 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 1.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 9.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.1 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 1.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 2.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 1.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 3.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.7 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.6 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 6.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.8 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 1.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 2.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.9 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.7 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 9.4 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 10.1 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 2.8 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 14.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 1.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 16.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 4.8 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.3 | 11.8 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.2 | 3.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 2.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 4.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 2.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.2 | 2.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.2 | 7.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 8.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 1.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 0.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 4.2 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 2.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 0.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 3.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 3.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 21.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.8 | 2.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.5 | 3.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.4 | 5.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.4 | 2.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.3 | 16.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 2.8 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.2 | 8.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 2.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 0.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 4.5 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.2 | 2.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 2.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 2.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 2.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 0.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 1.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 7.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 4.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 5.0 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.7 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |