Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
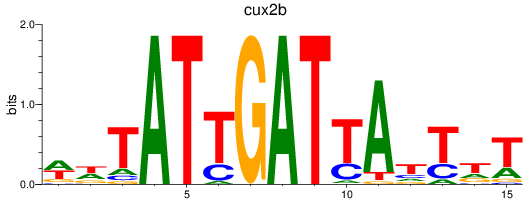
Results for cux2b
Z-value: 1.31
Transcription factors associated with cux2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cux2b
|
ENSDARG00000086345 | cut-like homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cux2b | dr11_v1_chr8_-_4097722_4097722 | -0.35 | 4.9e-04 | Click! |
Activity profile of cux2b motif
Sorted Z-values of cux2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_31264155 | 16.94 |
ENSDART00000012256
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr3_-_61205711 | 16.32 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr25_+_29160102 | 14.99 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr5_-_71705191 | 14.12 |
ENSDART00000187767
|
ak1
|
adenylate kinase 1 |
| chr11_-_18253111 | 13.76 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr25_+_31267268 | 13.09 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr11_+_11200550 | 13.02 |
ENSDART00000181339
ENSDART00000187116 |
myom2a
|
myomesin 2a |
| chr24_+_38301080 | 12.38 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr2_-_15324837 | 11.97 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr13_+_50375800 | 11.93 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr11_+_36243774 | 11.70 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr5_+_51848756 | 10.49 |
ENSDART00000087467
ENSDART00000184466 |
cmya5
|
cardiomyopathy associated 5 |
| chr19_+_43119698 | 10.38 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr23_+_32039386 | 8.84 |
ENSDART00000133801
|
mylk2
|
myosin light chain kinase 2 |
| chr6_+_3373665 | 8.79 |
ENSDART00000134133
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr19_-_48039400 | 8.32 |
ENSDART00000166748
ENSDART00000165921 |
csf3b
|
colony stimulating factor 3 (granulocyte) b |
| chr3_-_21242460 | 8.29 |
ENSDART00000007293
|
tcap
|
titin-cap (telethonin) |
| chr5_-_37886063 | 7.90 |
ENSDART00000131378
ENSDART00000132152 |
si:ch211-139a5.9
|
si:ch211-139a5.9 |
| chr2_-_30324610 | 7.85 |
ENSDART00000185422
|
jph1b
|
junctophilin 1b |
| chr20_+_20499869 | 7.72 |
ENSDART00000036124
|
six1b
|
SIX homeobox 1b |
| chr11_-_34065718 | 7.58 |
ENSDART00000110608
|
col6a1
|
collagen, type VI, alpha 1 |
| chr23_-_21515182 | 7.47 |
ENSDART00000142000
|
rnf207b
|
ring finger protein 207b |
| chr5_-_38820046 | 7.36 |
ENSDART00000182886
|
cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr8_+_554531 | 7.27 |
ENSDART00000193623
|
FO704758.2
|
|
| chr8_+_27807266 | 7.27 |
ENSDART00000170037
|
capza1b
|
capping protein (actin filament) muscle Z-line, alpha 1b |
| chr4_-_77135340 | 7.16 |
ENSDART00000180581
ENSDART00000179901 |
CU467646.7
|
|
| chr15_-_20933574 | 7.14 |
ENSDART00000152648
ENSDART00000152448 ENSDART00000152244 |
usp2a
|
ubiquitin specific peptidase 2a |
| chr1_-_45049603 | 7.13 |
ENSDART00000023336
|
rps6
|
ribosomal protein S6 |
| chr4_-_10599062 | 7.03 |
ENSDART00000048003
|
tspan12
|
tetraspanin 12 |
| chr23_-_31506854 | 6.88 |
ENSDART00000131352
ENSDART00000138625 ENSDART00000133002 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr2_+_51028269 | 6.69 |
ENSDART00000161254
|
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr22_+_396840 | 6.66 |
ENSDART00000163198
|
capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr14_+_46020571 | 6.53 |
ENSDART00000157617
ENSDART00000083928 |
c1qtnf2
|
C1q and TNF related 2 |
| chr16_+_54780544 | 6.42 |
ENSDART00000126646
|
si:zfos-1192g2.3
|
si:zfos-1192g2.3 |
| chr12_+_20641471 | 6.40 |
ENSDART00000133654
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr24_-_21952164 | 6.38 |
ENSDART00000058948
|
acot9.2
|
acyl-CoA thioesterase 9, tandem duplicate 2 |
| chr14_-_970853 | 6.37 |
ENSDART00000130801
|
acsl1b
|
acyl-CoA synthetase long chain family member 1b |
| chr10_-_7756865 | 6.20 |
ENSDART00000114373
ENSDART00000125407 ENSDART00000016317 |
loxa
|
lysyl oxidase a |
| chr17_-_2578026 | 6.19 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr16_-_33001153 | 6.16 |
ENSDART00000147941
|
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr12_-_49151326 | 6.01 |
ENSDART00000153244
|
bub3
|
BUB3 mitotic checkpoint protein |
| chr8_+_44783424 | 6.01 |
ENSDART00000025875
|
si:ch1073-459j12.1
|
si:ch1073-459j12.1 |
| chr17_-_2584423 | 5.98 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr19_+_48102560 | 5.85 |
ENSDART00000164464
|
utp18
|
UTP18 small subunit (SSU) processome component |
| chr17_+_27434626 | 5.75 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr14_-_899170 | 5.63 |
ENSDART00000165211
ENSDART00000031992 |
rgs14a
|
regulator of G protein signaling 14a |
| chr13_-_22843562 | 5.59 |
ENSDART00000142738
|
pbld
|
phenazine biosynthesis like protein domain containing |
| chr25_+_16194450 | 5.49 |
ENSDART00000141994
|
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr12_-_26407092 | 5.42 |
ENSDART00000178687
|
myoz1b
|
myozenin 1b |
| chr4_+_77933084 | 5.36 |
ENSDART00000148728
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr17_-_2573021 | 5.32 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr3_-_48559906 | 5.29 |
ENSDART00000160370
|
atp5pd
|
ATP synthase peripheral stalk subunit d |
| chr19_+_5674907 | 5.16 |
ENSDART00000042189
|
pdk2b
|
pyruvate dehydrogenase kinase, isozyme 2b |
| chr18_+_33468099 | 5.11 |
ENSDART00000131769
|
v2rh9
|
vomeronasal 2 receptor, h9 |
| chr12_-_48970299 | 5.01 |
ENSDART00000163734
|
rgrb
|
retinal G protein coupled receptor b |
| chr4_-_21652812 | 4.96 |
ENSDART00000174400
|
rps16
|
ribosomal protein S16 |
| chr25_+_37435720 | 4.95 |
ENSDART00000164390
|
chmp1a
|
charged multivesicular body protein 1A |
| chr13_+_35635672 | 4.92 |
ENSDART00000148481
|
thbs2a
|
thrombospondin 2a |
| chr17_-_8899323 | 4.88 |
ENSDART00000081590
|
nkl.1
|
NK-lysin tandem duplicate 1 |
| chr11_-_29658396 | 4.80 |
ENSDART00000183947
|
rpl22
|
ribosomal protein L22 |
| chr4_-_77135076 | 4.78 |
ENSDART00000174184
|
zgc:173770
|
zgc:173770 |
| chr19_-_38539670 | 4.76 |
ENSDART00000136775
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr4_-_12914163 | 4.69 |
ENSDART00000140002
ENSDART00000145917 ENSDART00000141355 ENSDART00000067135 |
msrb3
|
methionine sulfoxide reductase B3 |
| chr23_-_9925568 | 4.68 |
ENSDART00000081268
|
si:ch211-220i18.4
|
si:ch211-220i18.4 |
| chr23_+_32101202 | 4.63 |
ENSDART00000000992
|
zgc:56699
|
zgc:56699 |
| chr22_-_15602760 | 4.62 |
ENSDART00000009054
|
tpm4a
|
tropomyosin 4a |
| chr9_-_49493305 | 4.62 |
ENSDART00000148707
ENSDART00000148561 |
xirp2b
|
xin actin binding repeat containing 2b |
| chr1_+_27690 | 4.61 |
ENSDART00000162928
|
eed
|
embryonic ectoderm development |
| chr16_+_29509133 | 4.59 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr17_-_2590222 | 4.52 |
ENSDART00000185711
|
CR759892.1
|
|
| chr6_-_46742455 | 4.50 |
ENSDART00000011970
|
zgc:66479
|
zgc:66479 |
| chr18_+_16986495 | 4.50 |
ENSDART00000147377
|
si:ch211-218c6.8
|
si:ch211-218c6.8 |
| chr4_-_77125693 | 4.49 |
ENSDART00000174256
|
CU467646.3
|
|
| chr22_+_19290199 | 4.49 |
ENSDART00000148173
|
si:dkey-21e2.15
|
si:dkey-21e2.15 |
| chr18_-_20869175 | 4.42 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
| chr19_-_48391415 | 4.35 |
ENSDART00000170726
ENSDART00000169577 |
si:ch73-359m17.2
|
si:ch73-359m17.2 |
| chr2_-_42958619 | 4.32 |
ENSDART00000144317
|
oc90
|
otoconin 90 |
| chr16_+_35870456 | 4.22 |
ENSDART00000184321
|
thrap3a
|
thyroid hormone receptor associated protein 3a |
| chr14_-_52480661 | 4.19 |
ENSDART00000158353
|
exosc3
|
exosome component 3 |
| chr12_+_47663419 | 4.17 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr19_+_627899 | 4.13 |
ENSDART00000148508
|
tert
|
telomerase reverse transcriptase |
| chr14_+_46020282 | 4.11 |
ENSDART00000190087
|
c1qtnf2
|
C1q and TNF related 2 |
| chr2_+_20605186 | 4.08 |
ENSDART00000128505
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr6_+_2097690 | 4.07 |
ENSDART00000193770
|
tgm2b
|
transglutaminase 2b |
| chr4_+_25223050 | 4.05 |
ENSDART00000138121
|
itih5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr4_-_44500201 | 4.05 |
ENSDART00000150460
|
si:dkeyp-100h4.7
|
si:dkeyp-100h4.7 |
| chr11_-_29657947 | 4.02 |
ENSDART00000125753
|
rpl22
|
ribosomal protein L22 |
| chr9_+_9112943 | 4.00 |
ENSDART00000083033
|
sik1
|
salt-inducible kinase 1 |
| chr3_-_8272632 | 4.00 |
ENSDART00000143522
|
cyp2k6
|
cytochrome P450, family 2, subfamily K, polypeptide 6 |
| chr4_-_77130289 | 3.99 |
ENSDART00000174380
|
CU467646.7
|
|
| chr7_+_61480296 | 3.97 |
ENSDART00000083255
|
adam19a
|
ADAM metallopeptidase domain 19a |
| chr7_+_30933867 | 3.93 |
ENSDART00000156325
|
tjp1a
|
tight junction protein 1a |
| chr16_+_13965923 | 3.86 |
ENSDART00000103857
|
zgc:162509
|
zgc:162509 |
| chr14_+_30568961 | 3.84 |
ENSDART00000184303
|
mrpl11
|
mitochondrial ribosomal protein L11 |
| chr20_-_40754794 | 3.83 |
ENSDART00000187251
|
cx32.3
|
connexin 32.3 |
| chr2_+_45511459 | 3.83 |
ENSDART00000132101
|
aknad1
|
AKNA domain containing 1 |
| chr25_+_37290206 | 3.83 |
ENSDART00000086474
|
si:dkey-234i14.12
|
si:dkey-234i14.12 |
| chr13_-_290377 | 3.79 |
ENSDART00000134963
|
chs1
|
chitin synthase 1 |
| chr10_-_21545091 | 3.76 |
ENSDART00000029122
ENSDART00000132207 |
zgc:165539
|
zgc:165539 |
| chr9_+_13120419 | 3.75 |
ENSDART00000141005
|
fam117bb
|
family with sequence similarity 117, member Bb |
| chr13_+_46941930 | 3.71 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr10_-_24391716 | 3.69 |
ENSDART00000141332
ENSDART00000100772 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr7_+_30626378 | 3.68 |
ENSDART00000173533
ENSDART00000052541 |
ccnb2
|
cyclin B2 |
| chr4_-_77140030 | 3.67 |
ENSDART00000174332
ENSDART00000190394 ENSDART00000189208 |
CU467646.5
CU467646.7
|
|
| chr9_-_49964810 | 3.67 |
ENSDART00000167098
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr14_-_34512859 | 3.61 |
ENSDART00000140368
|
si:ch211-232m8.3
|
si:ch211-232m8.3 |
| chr7_-_4461104 | 3.59 |
ENSDART00000023090
ENSDART00000140770 |
slc12a10.1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 1 |
| chr2_+_24936766 | 3.58 |
ENSDART00000025962
|
gyg1a
|
glycogenin 1a |
| chr1_+_57041549 | 3.54 |
ENSDART00000152198
|
si:ch211-1f22.16
|
si:ch211-1f22.16 |
| chr11_-_37589293 | 3.54 |
ENSDART00000172989
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr24_-_31306724 | 3.53 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr5_+_6672870 | 3.51 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr2_+_20605925 | 3.49 |
ENSDART00000191510
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr10_-_40964739 | 3.46 |
ENSDART00000076359
|
npffr1l1
|
neuropeptide FF receptor 1 like 1 |
| chr1_-_38815361 | 3.44 |
ENSDART00000148790
ENSDART00000148572 ENSDART00000149080 |
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr13_-_9442942 | 3.43 |
ENSDART00000138833
|
grxcr1
|
glutaredoxin, cysteine rich 1 |
| chr12_+_16284086 | 3.40 |
ENSDART00000013360
ENSDART00000141169 |
ppp1r3cb
|
protein phosphatase 1, regulatory subunit 3Cb |
| chr25_+_22320738 | 3.38 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr25_-_37319117 | 3.36 |
ENSDART00000044499
|
igl4v9
|
immunoglobulin light 4 variable 9 |
| chr19_-_40199081 | 3.36 |
ENSDART00000051970
ENSDART00000151079 |
grn2
|
granulin 2 |
| chr21_-_19314618 | 3.36 |
ENSDART00000188744
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr23_-_33750135 | 3.33 |
ENSDART00000187641
|
bin2a
|
bridging integrator 2a |
| chr9_-_23990416 | 3.28 |
ENSDART00000113176
|
col6a3
|
collagen, type VI, alpha 3 |
| chr21_+_17768174 | 3.28 |
ENSDART00000141380
|
rxraa
|
retinoid X receptor, alpha a |
| chr2_-_45510699 | 3.26 |
ENSDART00000024034
ENSDART00000145634 |
gpsm2
|
G protein signaling modulator 2 |
| chr13_-_27354003 | 3.21 |
ENSDART00000101479
ENSDART00000044652 |
ddx43
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 |
| chr25_+_37297659 | 3.20 |
ENSDART00000086733
|
si:dkey-234i14.13
|
si:dkey-234i14.13 |
| chr24_+_25913162 | 3.17 |
ENSDART00000143099
ENSDART00000184814 |
map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr7_+_4824882 | 3.17 |
ENSDART00000137369
|
si:dkey-28d5.7
|
si:dkey-28d5.7 |
| chr8_-_4327473 | 3.17 |
ENSDART00000134378
|
cux2b
|
cut-like homeobox 2b |
| chr7_-_38658411 | 3.16 |
ENSDART00000109463
ENSDART00000017155 |
npsn
|
nephrosin |
| chr11_-_27962757 | 3.13 |
ENSDART00000147386
|
ece1
|
endothelin converting enzyme 1 |
| chr6_+_52235441 | 3.13 |
ENSDART00000056319
|
cox6c
|
cytochrome c oxidase subunit VIc |
| chr4_+_44454270 | 3.12 |
ENSDART00000150482
|
si:dkeyp-100h4.4
|
si:dkeyp-100h4.4 |
| chr18_-_1414760 | 3.12 |
ENSDART00000171881
|
PEPD
|
peptidase D |
| chr16_-_13965518 | 3.08 |
ENSDART00000138304
|
si:dkey-85k15.4
|
si:dkey-85k15.4 |
| chr3_-_5277041 | 3.04 |
ENSDART00000046995
|
txn2
|
thioredoxin 2 |
| chr24_-_39826865 | 3.03 |
ENSDART00000089232
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr20_-_23842631 | 3.02 |
ENSDART00000153079
|
si:dkey-15j16.3
|
si:dkey-15j16.3 |
| chr6_-_33875919 | 2.99 |
ENSDART00000190411
|
tmem69
|
transmembrane protein 69 |
| chr11_+_10984293 | 2.98 |
ENSDART00000065933
|
itgb6
|
integrin, beta 6 |
| chr25_+_36349574 | 2.98 |
ENSDART00000184101
|
zgc:173552
|
zgc:173552 |
| chr17_-_15747296 | 2.96 |
ENSDART00000153970
|
cx52.7
|
connexin 52.7 |
| chr1_-_34713710 | 2.93 |
ENSDART00000141569
ENSDART00000102111 ENSDART00000122137 |
tpt1
|
tumor protein, translationally-controlled 1 |
| chr17_-_681142 | 2.93 |
ENSDART00000165583
|
soul3
|
heme-binding protein soul3 |
| chr7_-_6441865 | 2.92 |
ENSDART00000172831
|
hist1h2a10
|
histone cluster 1 H2A family member 10 |
| chr8_-_3773374 | 2.92 |
ENSDART00000115036
|
bicdl1
|
BICD family like cargo adaptor 1 |
| chr20_+_38910214 | 2.91 |
ENSDART00000047362
|
msra
|
methionine sulfoxide reductase A |
| chr17_-_34952562 | 2.90 |
ENSDART00000021128
|
kidins220a
|
kinase D-interacting substrate 220a |
| chr11_+_37201483 | 2.90 |
ENSDART00000160930
ENSDART00000173439 ENSDART00000171273 |
zgc:112265
|
zgc:112265 |
| chr9_-_5318873 | 2.88 |
ENSDART00000129308
|
ACVR1C
|
activin A receptor type 1C |
| chr25_-_19661198 | 2.88 |
ENSDART00000149641
|
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr21_-_5881344 | 2.86 |
ENSDART00000009241
|
rpl35
|
ribosomal protein L35 |
| chr2_+_36004381 | 2.86 |
ENSDART00000098706
|
lamc2
|
laminin, gamma 2 |
| chr8_-_45760087 | 2.86 |
ENSDART00000025620
|
ppiaa
|
peptidylprolyl isomerase Aa (cyclophilin A) |
| chr19_+_34274504 | 2.84 |
ENSDART00000132046
|
si:ch211-9n13.3
|
si:ch211-9n13.3 |
| chr2_+_21634128 | 2.82 |
ENSDART00000089822
|
fbxl7
|
F-box and leucine-rich repeat protein 7 |
| chr20_+_20726231 | 2.82 |
ENSDART00000147112
|
zgc:193541
|
zgc:193541 |
| chr21_+_5531138 | 2.82 |
ENSDART00000163825
|
ly6m6
|
lymphocyte antigen 6 family member M6 |
| chr3_-_8246309 | 2.81 |
ENSDART00000134497
ENSDART00000015232 |
cyp2k22
|
cytochrome P450, family 2, subfamily K, polypeptide 22 |
| chr23_-_43424510 | 2.80 |
ENSDART00000055564
|
rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr13_+_51869025 | 2.80 |
ENSDART00000187066
|
LT631684.1
|
|
| chr23_-_5818992 | 2.79 |
ENSDART00000148730
|
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr4_-_17680493 | 2.77 |
ENSDART00000180131
|
BX890572.2
|
|
| chr17_+_35362851 | 2.76 |
ENSDART00000137659
|
cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr9_+_21990095 | 2.76 |
ENSDART00000146829
ENSDART00000133515 ENSDART00000193582 |
si:dkey-57a22.13
|
si:dkey-57a22.13 |
| chr4_+_25574827 | 2.75 |
ENSDART00000187726
|
zgc:195175
|
zgc:195175 |
| chr7_-_73854476 | 2.75 |
ENSDART00000186481
|
zgc:173552
|
zgc:173552 |
| chr9_+_55455801 | 2.74 |
ENSDART00000144757
ENSDART00000186543 |
mxra5b
|
matrix-remodelling associated 5b |
| chr1_-_52498146 | 2.72 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr18_-_6766354 | 2.71 |
ENSDART00000132611
|
adm2b
|
adrenomedullin 2b |
| chr13_+_37273010 | 2.69 |
ENSDART00000144387
|
gphb5
|
glycoprotein hormone beta 5 |
| chr12_+_9342502 | 2.66 |
ENSDART00000152808
|
kcnh6b
|
potassium voltage-gated channel, subfamily H (eag-related), member 6b |
| chr7_-_29292206 | 2.66 |
ENSDART00000086753
|
dapk2a
|
death-associated protein kinase 2a |
| chr13_+_37273221 | 2.65 |
ENSDART00000035011
|
gphb5
|
glycoprotein hormone beta 5 |
| chr2_+_56657804 | 2.64 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr2_-_19520324 | 2.62 |
ENSDART00000079877
|
pimr52
|
Pim proto-oncogene, serine/threonine kinase, related 52 |
| chr23_-_45705525 | 2.62 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr2_-_6262441 | 2.60 |
ENSDART00000092190
|
arl14
|
ADP-ribosylation factor-like 14 |
| chr8_+_3496204 | 2.58 |
ENSDART00000085993
|
pxnb
|
paxillin b |
| chr11_+_21096339 | 2.58 |
ENSDART00000124574
|
il19l
|
interleukin 19 like |
| chr2_-_51794472 | 2.58 |
ENSDART00000186652
|
BX908782.3
|
|
| chr4_-_9852318 | 2.58 |
ENSDART00000080702
|
glt8d2
|
glycosyltransferase 8 domain containing 2 |
| chr2_-_7845110 | 2.57 |
ENSDART00000091987
|
si:ch211-38m6.7
|
si:ch211-38m6.7 |
| chr8_+_27807974 | 2.56 |
ENSDART00000078509
|
capza1b
|
capping protein (actin filament) muscle Z-line, alpha 1b |
| chr1_+_524717 | 2.56 |
ENSDART00000102421
ENSDART00000184473 |
mrpl16
|
mitochondrial ribosomal protein L16 |
| chr1_+_292545 | 2.53 |
ENSDART00000148261
|
cenpe
|
centromere protein E |
| chr24_-_27452488 | 2.52 |
ENSDART00000136433
|
ccl34b.8
|
chemokine (C-C motif) ligand 34b, duplicate 8 |
| chr7_-_35432901 | 2.52 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr5_+_56026031 | 2.51 |
ENSDART00000050970
|
fzd9a
|
frizzled class receptor 9a |
| chr3_-_34096987 | 2.51 |
ENSDART00000151663
|
ighv4-5
|
immunoglobulin heavy variable 4-5 |
| chr12_+_47698356 | 2.51 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr25_-_37322081 | 2.50 |
ENSDART00000128285
|
igl4v10
|
immunoglobulin light 4 variable 10 |
| chr8_-_53044300 | 2.49 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr12_+_16168342 | 2.49 |
ENSDART00000079326
ENSDART00000170024 |
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr17_+_53435279 | 2.47 |
ENSDART00000126630
|
cx52.9
|
connexin 52.9 |
| chr17_+_6276559 | 2.47 |
ENSDART00000131075
|
dusp23b
|
dual specificity phosphatase 23b |
| chr13_+_12671513 | 2.47 |
ENSDART00000010517
|
eif4eb
|
eukaryotic translation initiation factor 4eb |
| chr7_+_16991711 | 2.47 |
ENSDART00000173660
|
nav2a
|
neuron navigator 2a |
| chr10_-_41352502 | 2.46 |
ENSDART00000052971
ENSDART00000128156 |
rab11fip1b
|
RAB11 family interacting protein 1 (class I) b |
Network of associatons between targets according to the STRING database.
First level regulatory network of cux2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 13.8 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 2.6 | 7.7 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 2.5 | 7.5 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 2.1 | 6.4 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 2.1 | 8.3 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 2.0 | 14.1 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 2.0 | 5.9 | GO:1901376 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 1.8 | 5.3 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 1.7 | 6.9 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 1.7 | 17.1 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 1.4 | 4.2 | GO:0071034 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 1.4 | 4.1 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 1.3 | 6.7 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 1.3 | 3.9 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 1.2 | 4.7 | GO:0030091 | protein repair(GO:0030091) |
| 1.0 | 17.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 1.0 | 5.1 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.9 | 5.7 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.9 | 3.7 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.9 | 2.8 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.9 | 11.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.9 | 5.5 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.8 | 6.9 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.8 | 3.8 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.7 | 4.5 | GO:0032648 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.7 | 2.9 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.7 | 7.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.7 | 2.1 | GO:0099623 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.7 | 2.1 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 0.7 | 7.5 | GO:1902547 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.7 | 3.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.7 | 2.0 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.6 | 3.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.6 | 2.4 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.6 | 30.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.6 | 1.8 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.6 | 2.4 | GO:0070257 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.6 | 2.8 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.5 | 2.1 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.5 | 1.6 | GO:0006565 | L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 0.5 | 2.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.5 | 1.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.5 | 16.8 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.5 | 2.4 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.5 | 8.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.4 | 1.3 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.4 | 8.0 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.4 | 1.8 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.4 | 1.8 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.4 | 2.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.4 | 2.2 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.4 | 1.3 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.4 | 2.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.4 | 20.0 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.4 | 3.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.4 | 6.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.4 | 1.6 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.4 | 2.6 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.4 | 2.6 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.4 | 1.9 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) positive regulation of reactive oxygen species metabolic process(GO:2000379) |
| 0.4 | 6.2 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.4 | 3.9 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.4 | 3.5 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.3 | 1.7 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.3 | 1.7 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.3 | 2.4 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.3 | 1.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.3 | 5.6 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.3 | 4.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.3 | 0.9 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.3 | 2.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 4.3 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.3 | 7.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 0.9 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.3 | 6.8 | GO:0071174 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.3 | 2.3 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.3 | 0.8 | GO:0002456 | T cell cytokine production(GO:0002369) T cell mediated immunity(GO:0002456) |
| 0.3 | 3.0 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.3 | 0.8 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.3 | 1.9 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.3 | 1.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.3 | 2.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.3 | 17.1 | GO:0006414 | translational elongation(GO:0006414) |
| 0.3 | 1.6 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.3 | 3.6 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.3 | 1.5 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.3 | 15.8 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.2 | 3.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.2 | 6.6 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.2 | 0.7 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.2 | 1.2 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 | 1.2 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.2 | 9.0 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.2 | 2.2 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.2 | 1.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.2 | 4.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.2 | 3.1 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 9.1 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.2 | 2.2 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.2 | 0.8 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.2 | 0.9 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 0.6 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.2 | 4.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 1.3 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.2 | 1.3 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.2 | 2.6 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.2 | 3.9 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.2 | 0.5 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 0.5 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.2 | 5.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.2 | 2.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.2 | 0.5 | GO:0036314 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.2 | 6.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.2 | 1.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 1.8 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.2 | 5.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.2 | 4.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 1.7 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.2 | 1.8 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 1.0 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 3.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.7 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 7.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 1.0 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 4.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.5 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 6.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.8 | GO:0031960 | response to corticosteroid(GO:0031960) response to glucocorticoid(GO:0051384) |
| 0.1 | 0.5 | GO:0055130 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.1 | 4.9 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 1.5 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 1.3 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 2.6 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.1 | 0.9 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 1.6 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 3.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.9 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.6 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 0.7 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 0.7 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 1.7 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.1 | 2.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.7 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 1.1 | GO:2000403 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.1 | 0.5 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 0.4 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.1 | 0.8 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 | 0.8 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.1 | 4.7 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 0.7 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 1.2 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 0.8 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.8 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.6 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 1.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 2.8 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.1 | 2.4 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.1 | 3.2 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.1 | 0.6 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 2.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.3 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 1.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 1.2 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 1.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 7.7 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 1.6 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 4.2 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.1 | 0.3 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 1.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 2.9 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.1 | 1.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 5.1 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 1.8 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.1 | 3.1 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.1 | 0.5 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 1.4 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 18.1 | GO:0060537 | muscle tissue development(GO:0060537) |
| 0.1 | 7.3 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 1.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 1.2 | GO:2001236 | regulation of extrinsic apoptotic signaling pathway(GO:2001236) |
| 0.1 | 7.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 5.8 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 0.9 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 2.7 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 1.6 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 1.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 1.1 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 4.3 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 1.9 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 2.3 | GO:0043627 | response to estrogen(GO:0043627) cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.6 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 1.2 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 1.3 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 1.9 | GO:0009566 | single fertilization(GO:0007338) fertilization(GO:0009566) |
| 0.0 | 1.6 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 1.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.5 | GO:0006030 | aminoglycan catabolic process(GO:0006026) chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) glucosamine-containing compound metabolic process(GO:1901071) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 0.5 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.5 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 1.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 2.2 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 1.2 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.3 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 3.4 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.4 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.9 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.3 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 1.0 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 1.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.9 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 1.5 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 1.4 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.0 | 1.0 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.1 | GO:0060911 | cardiac cell fate commitment(GO:0060911) cardiac cell fate specification(GO:0060912) |
| 0.0 | 1.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 3.5 | GO:0001503 | ossification(GO:0001503) |
| 0.0 | 12.6 | GO:0007346 | regulation of mitotic cell cycle(GO:0007346) |
| 0.0 | 1.3 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.9 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 1.5 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 6.4 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 1.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 1.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.4 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) amino acid activation(GO:0043038) tRNA aminoacylation(GO:0043039) |
| 0.0 | 2.7 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 1.4 | GO:0009190 | cyclic nucleotide biosynthetic process(GO:0009190) |
| 0.0 | 6.4 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.7 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 1.5 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.0 | 0.8 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 1.3 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.1 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 2.5 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 5.2 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 0.4 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 | 0.2 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.7 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.9 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 1.0 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 16.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 1.6 | 7.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 1.4 | 4.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 1.0 | 7.0 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 1.0 | 3.0 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 1.0 | 6.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.8 | 5.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.8 | 2.4 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.7 | 30.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.6 | 13.0 | GO:0031430 | M band(GO:0031430) |
| 0.6 | 8.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.6 | 2.3 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.5 | 7.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.5 | 3.8 | GO:0030428 | cell septum(GO:0030428) |
| 0.4 | 1.7 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.4 | 2.0 | GO:0034657 | GID complex(GO:0034657) |
| 0.4 | 17.1 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.4 | 4.2 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.4 | 3.5 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.3 | 4.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.3 | 4.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 1.5 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.3 | 4.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 2.0 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.3 | 1.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.3 | 1.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.3 | 6.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.3 | 0.8 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.2 | 3.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 2.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.2 | 8.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 18.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 1.9 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.2 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 3.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.2 | 1.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 1.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 26.1 | GO:0005840 | ribosome(GO:0005840) |
| 0.1 | 2.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 6.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 4.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 1.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 4.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.1 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 0.9 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 3.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.8 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 8.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.0 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 3.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 3.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 28.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 7.9 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.1 | 1.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 6.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 7.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 5.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 2.2 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.1 | 7.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 0.8 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 3.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 12.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 1.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 1.2 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 4.1 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.4 | GO:0005903 | brush border(GO:0005903) brush border membrane(GO:0031526) |
| 0.0 | 10.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 4.9 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 63.0 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 1.1 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 1.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.0 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 7.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.7 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.6 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 17.9 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.8 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 15.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.8 | 7.2 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 1.6 | 17.5 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 1.4 | 2.8 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 1.4 | 4.1 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 1.3 | 8.8 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 1.1 | 3.4 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 1.1 | 14.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 1.1 | 6.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.9 | 5.5 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.9 | 6.9 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.8 | 2.4 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.8 | 4.7 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.8 | 0.8 | GO:0031543 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) peptidyl-proline dioxygenase activity(GO:0031543) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.7 | 5.7 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.7 | 3.5 | GO:0047192 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.7 | 8.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.6 | 5.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.6 | 3.8 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.6 | 1.7 | GO:0031835 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.6 | 6.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.5 | 5.4 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.5 | 15.1 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.5 | 8.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.5 | 1.6 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.5 | 3.5 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.5 | 3.8 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.4 | 3.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.4 | 2.2 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.4 | 5.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.4 | 1.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.4 | 4.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.4 | 3.6 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.4 | 11.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.4 | 1.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.4 | 2.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.4 | 1.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.4 | 17.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.4 | 2.6 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.4 | 6.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.4 | 3.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 2.9 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.3 | 2.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.3 | 1.7 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.3 | 4.9 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 5.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 1.8 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.3 | 2.1 | GO:0031779 | melanocortin receptor binding(GO:0031779) |
| 0.3 | 4.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 1.6 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.3 | 2.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.3 | 1.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.3 | 1.3 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.2 | 3.0 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.2 | 1.2 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 5.7 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 0.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 0.9 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.2 | 1.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 4.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 3.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.2 | 1.6 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.2 | 1.0 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.2 | 1.5 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.2 | 3.0 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.2 | 0.6 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 2.4 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 2.9 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 2.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 11.3 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.2 | 4.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.2 | 2.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.2 | 1.3 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.2 | 2.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 31.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 5.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 1.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 2.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.8 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 8.9 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 1.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 6.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 1.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 2.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 6.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 5.0 | GO:0019957 | chemokine binding(GO:0019956) C-C chemokine binding(GO:0019957) |
| 0.1 | 2.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.4 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 1.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.5 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 1.3 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 2.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 8.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.7 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 3.0 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.9 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 1.9 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.8 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 1.8 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 1.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 15.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 2.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 2.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 0.8 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 1.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.8 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 1.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 1.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 2.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 29.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 4.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 16.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 1.8 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 1.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.8 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 1.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 1.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 5.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.4 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 7.7 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 0.2 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.1 | 1.3 | GO:0008236 | serine-type peptidase activity(GO:0008236) serine hydrolase activity(GO:0017171) |
| 0.1 | 1.9 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 6.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 6.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 5.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 2.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.7 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.1 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.2 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 8.0 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 5.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 9.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 2.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.6 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.6 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 1.7 | GO:0004175 | endopeptidase activity(GO:0004175) |
| 0.0 | 1.0 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.3 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.7 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 1.1 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 2.1 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 3.5 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 1.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 8.9 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 1.3 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 2.7 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.0 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 1.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 14.8 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 14.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 1.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 3.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 15.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.4 | 14.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.4 | 4.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.4 | 8.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 12.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.3 | 3.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 2.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 13.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 9.2 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 3.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 2.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 3.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 1.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 0.7 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 1.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 5.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 3.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.4 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 14.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.4 | 3.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.4 | 3.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 4.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.4 | 4.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.4 | 3.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.4 | 1.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.3 | 2.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 5.8 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.3 | 4.7 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.3 | 15.6 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.3 | 8.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 6.0 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.2 | 12.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 3.7 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 1.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 1.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 3.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 2.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 1.8 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 11.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 2.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 2.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 3.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 1.7 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 0.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 0.8 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 1.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 0.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 2.2 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 0.9 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 5.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 0.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 4.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 2.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 2.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.6 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 1.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.1 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 2.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 2.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |