Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
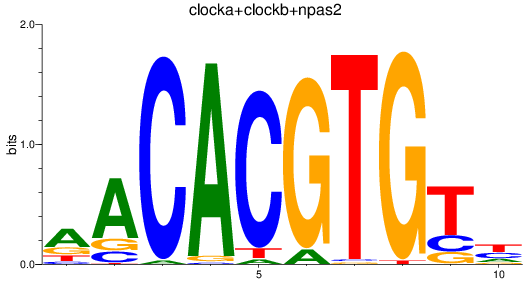
Results for clocka+clockb+npas2
Z-value: 1.88
Transcription factors associated with clocka+clockb+npas2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
clockb
|
ENSDARG00000003631 | clock circadian regulator b |
|
clocka
|
ENSDARG00000011703 | clock circadian regulator a |
|
npas2
|
ENSDARG00000016536 | neuronal PAS domain protein 2 |
|
npas2
|
ENSDARG00000116993 | neuronal PAS domain protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| clocka | dr11_v1_chr20_+_22067337_22067337 | -0.66 | 5.7e-13 | Click! |
| clockb | dr11_v1_chr1_+_19434198_19434198 | -0.49 | 4.9e-07 | Click! |
| npas2 | dr11_v1_chr5_+_22791686_22791686 | -0.37 | 2.6e-04 | Click! |
Activity profile of clocka+clockb+npas2 motif
Sorted Z-values of clocka+clockb+npas2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_43119698 | 22.88 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr14_+_22076596 | 22.76 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr21_+_11969603 | 21.18 |
ENSDART00000142247
ENSDART00000140652 |
mlnl
|
motilin-like |
| chr10_+_22034477 | 18.76 |
ENSDART00000133304
ENSDART00000134189 ENSDART00000021240 ENSDART00000100526 |
npm1a
|
nucleophosmin 1a |
| chr19_-_47555956 | 17.76 |
ENSDART00000114549
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr20_-_29483514 | 17.13 |
ENSDART00000062370
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr24_-_12938922 | 16.26 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr22_-_23612854 | 11.93 |
ENSDART00000165885
|
cfhl5
|
complement factor H like 5 |
| chr20_+_10544100 | 11.78 |
ENSDART00000113927
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr16_-_26537103 | 11.73 |
ENSDART00000134908
ENSDART00000008152 |
sgk2b
|
serum/glucocorticoid regulated kinase 2b |
| chr21_-_20765338 | 11.66 |
ENSDART00000135940
|
ghrb
|
growth hormone receptor b |
| chr9_+_20781047 | 11.33 |
ENSDART00000139174
|
fam46c
|
family with sequence similarity 46, member C |
| chr16_-_31919568 | 10.87 |
ENSDART00000027364
|
rbfox1l
|
RNA binding fox-1 homolog 1, like |
| chr11_-_16975190 | 10.63 |
ENSDART00000122222
|
suclg2
|
succinate-CoA ligase, GDP-forming, beta subunit |
| chr10_+_17026870 | 10.59 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr3_-_18805225 | 9.67 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr10_-_322769 | 9.58 |
ENSDART00000165244
|
akt2l
|
v-akt murine thymoma viral oncogene homolog 2, like |
| chr5_+_38462121 | 9.38 |
ENSDART00000144425
|
gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr23_+_25292147 | 9.35 |
ENSDART00000131486
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr5_-_1963498 | 9.19 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr7_+_6652967 | 9.18 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr2_-_42375275 | 9.14 |
ENSDART00000026339
|
gtpbp4
|
GTP binding protein 4 |
| chr5_+_19337108 | 9.10 |
ENSDART00000089078
|
acacb
|
acetyl-CoA carboxylase beta |
| chr3_-_16760923 | 9.06 |
ENSDART00000055855
|
aspdh
|
aspartate dehydrogenase domain containing |
| chr6_+_50451337 | 9.01 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr17_-_49407091 | 8.87 |
ENSDART00000021950
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr24_-_42090635 | 8.67 |
ENSDART00000166413
|
ssr1
|
signal sequence receptor, alpha |
| chr6_-_8498908 | 8.57 |
ENSDART00000149222
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr11_-_29946927 | 8.50 |
ENSDART00000165182
|
zgc:113276
|
zgc:113276 |
| chr14_+_14836468 | 8.46 |
ENSDART00000166728
|
si:dkey-102m7.3
|
si:dkey-102m7.3 |
| chr18_-_19456269 | 8.30 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr7_+_67467702 | 8.27 |
ENSDART00000168460
ENSDART00000170322 |
rpl13
|
ribosomal protein L13 |
| chr8_+_26059677 | 8.25 |
ENSDART00000009178
|
impdh2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr11_-_28050559 | 8.13 |
ENSDART00000136859
|
ece1
|
endothelin converting enzyme 1 |
| chr11_-_29946640 | 8.11 |
ENSDART00000079175
|
zgc:113276
|
zgc:113276 |
| chr20_-_49704915 | 8.01 |
ENSDART00000189232
|
COX7A2 (1 of many)
|
cytochrome c oxidase subunit 7A2 |
| chr3_-_58798377 | 8.01 |
ENSDART00000161248
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr18_+_15644559 | 7.97 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr13_-_31370184 | 7.78 |
ENSDART00000034829
|
rrp12
|
ribosomal RNA processing 12 homolog |
| chr11_-_37995501 | 7.65 |
ENSDART00000192096
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr22_-_20695237 | 7.62 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr12_+_48390715 | 7.61 |
ENSDART00000149351
|
scd
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr13_-_42306348 | 7.61 |
ENSDART00000003706
|
kmo
|
kynurenine 3-monooxygenase |
| chr21_+_43702016 | 7.57 |
ENSDART00000017176
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr22_+_835728 | 7.56 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr20_+_25626479 | 7.55 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr4_+_25607743 | 7.48 |
ENSDART00000028297
|
acot14
|
acyl-CoA thioesterase 14 |
| chr3_+_18398876 | 7.47 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr2_-_51630555 | 7.43 |
ENSDART00000171746
|
pigr
|
polymeric immunoglobulin receptor |
| chr2_+_37875789 | 7.32 |
ENSDART00000036318
ENSDART00000127679 |
cbln13
|
cerebellin 13 |
| chr24_+_34069675 | 7.28 |
ENSDART00000143995
|
si:ch211-190p8.2
|
si:ch211-190p8.2 |
| chr11_+_3959495 | 7.17 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr6_-_52348562 | 7.14 |
ENSDART00000142565
ENSDART00000145369 ENSDART00000016890 |
eif6
|
eukaryotic translation initiation factor 6 |
| chr9_-_33328948 | 7.12 |
ENSDART00000006948
|
rpl8
|
ribosomal protein L8 |
| chr15_+_32711663 | 7.08 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr11_+_29671661 | 7.00 |
ENSDART00000024318
ENSDART00000165024 |
rnf207a
|
ring finger protein 207a |
| chr7_-_38644560 | 6.96 |
ENSDART00000114934
|
c6ast3
|
six-cysteine containing astacin protease 3 |
| chr7_+_41812190 | 6.92 |
ENSDART00000113732
ENSDART00000174137 |
orc6
|
origin recognition complex, subunit 6 |
| chr8_-_21372446 | 6.91 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr4_-_16824231 | 6.89 |
ENSDART00000014007
|
gys2
|
glycogen synthase 2 |
| chr14_+_23518110 | 6.86 |
ENSDART00000112930
|
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr12_+_30586599 | 6.85 |
ENSDART00000124920
ENSDART00000126984 |
nrap
|
nebulin-related anchoring protein |
| chr4_-_16824556 | 6.84 |
ENSDART00000165289
ENSDART00000185839 |
gys2
|
glycogen synthase 2 |
| chr19_+_42983613 | 6.81 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr21_-_22724980 | 6.79 |
ENSDART00000035469
|
c1qa
|
complement component 1, q subcomponent, A chain |
| chr5_+_13870340 | 6.77 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr12_+_19384615 | 6.76 |
ENSDART00000078266
|
rsl1d1
|
ribosomal L1 domain containing 1 |
| chr5_+_72194444 | 6.73 |
ENSDART00000165436
|
ddx54
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 |
| chr8_+_23213320 | 6.71 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr19_+_32166702 | 6.70 |
ENSDART00000021798
|
fabp11a
|
fatty acid binding protein 11a |
| chr3_+_16229911 | 6.70 |
ENSDART00000121728
|
rpl19
|
ribosomal protein L19 |
| chr16_-_48400639 | 6.68 |
ENSDART00000159372
|
eif3ha
|
eukaryotic translation initiation factor 3, subunit H, a |
| chr17_+_53156530 | 6.66 |
ENSDART00000126277
ENSDART00000156774 |
dph6
|
diphthamine biosynthesis 6 |
| chr9_-_33329700 | 6.59 |
ENSDART00000147265
ENSDART00000140039 |
rpl8
|
ribosomal protein L8 |
| chr22_+_661711 | 6.56 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr3_-_58798815 | 6.50 |
ENSDART00000082920
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr6_+_29305190 | 6.42 |
ENSDART00000078647
|
si:ch211-201h21.5
|
si:ch211-201h21.5 |
| chr7_+_41812817 | 6.39 |
ENSDART00000174165
|
orc6
|
origin recognition complex, subunit 6 |
| chr3_-_3398383 | 6.36 |
ENSDART00000047865
|
si:dkey-46g23.2
|
si:dkey-46g23.2 |
| chr18_+_8917766 | 6.31 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr21_-_45382112 | 6.27 |
ENSDART00000151029
ENSDART00000151335 ENSDART00000151687 ENSDART00000075438 |
cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chr8_-_18203274 | 6.23 |
ENSDART00000134078
ENSDART00000180235 ENSDART00000080006 ENSDART00000125418 ENSDART00000142114 |
elovl8b
|
ELOVL fatty acid elongase 8b |
| chr15_+_25635326 | 6.15 |
ENSDART00000135409
ENSDART00000162240 ENSDART00000052645 |
tsr1
|
TSR1, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr7_+_41812636 | 6.09 |
ENSDART00000174333
|
orc6
|
origin recognition complex, subunit 6 |
| chr9_-_11587070 | 6.08 |
ENSDART00000030995
|
umps
|
uridine monophosphate synthetase |
| chr9_-_56231387 | 6.07 |
ENSDART00000149851
|
rpl31
|
ribosomal protein L31 |
| chr22_+_661505 | 6.07 |
ENSDART00000149460
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr9_+_20780813 | 6.07 |
ENSDART00000142787
|
fam46c
|
family with sequence similarity 46, member C |
| chr21_+_26071874 | 6.06 |
ENSDART00000003001
ENSDART00000146573 |
rpl23a
|
ribosomal protein L23a |
| chr8_-_18203092 | 6.00 |
ENSDART00000140620
|
elovl8b
|
ELOVL fatty acid elongase 8b |
| chr5_-_42060121 | 6.00 |
ENSDART00000148021
ENSDART00000147407 |
cenpv
|
centromere protein V |
| chr16_-_34195002 | 5.98 |
ENSDART00000054026
|
rcc1
|
regulator of chromosome condensation 1 |
| chr3_-_25377163 | 5.96 |
ENSDART00000055490
|
kpna2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr16_+_25245857 | 5.95 |
ENSDART00000155220
|
klhl38b
|
kelch-like family member 38b |
| chr16_-_13388821 | 5.93 |
ENSDART00000144062
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr22_+_29991834 | 5.91 |
ENSDART00000147728
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr2_+_25839650 | 5.90 |
ENSDART00000134077
ENSDART00000140804 |
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr5_-_32505109 | 5.90 |
ENSDART00000188219
|
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr12_+_47081783 | 5.89 |
ENSDART00000158568
|
mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr8_-_11229523 | 5.89 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr3_+_1150348 | 5.87 |
ENSDART00000148524
|
nol12
|
nucleolar protein 12 |
| chr10_+_17714866 | 5.86 |
ENSDART00000039969
|
slc20a1b
|
solute carrier family 20 (phosphate transporter), member 1b |
| chr25_+_28555584 | 5.86 |
ENSDART00000157046
|
SLC15A5
|
si:ch211-190o6.3 |
| chr14_+_94603 | 5.84 |
ENSDART00000162480
|
mcm7
|
minichromosome maintenance complex component 7 |
| chr7_-_38644287 | 5.83 |
ENSDART00000182307
|
c6ast3
|
six-cysteine containing astacin protease 3 |
| chr16_+_16969060 | 5.81 |
ENSDART00000182819
ENSDART00000191876 |
si:ch211-120k19.1
rpl18
|
si:ch211-120k19.1 ribosomal protein L18 |
| chr23_+_25291891 | 5.75 |
ENSDART00000016248
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr7_+_69841017 | 5.67 |
ENSDART00000169107
|
FO818704.1
|
|
| chr4_-_4570475 | 5.62 |
ENSDART00000184955
|
rassf3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr13_+_15701849 | 5.61 |
ENSDART00000003517
|
trmt61a
|
tRNA methyltransferase 61A |
| chr5_+_37517800 | 5.58 |
ENSDART00000048107
|
FP102018.1
|
Danio rerio latent transforming growth factor beta binding protein 3 (ltbp3), mRNA. |
| chr12_+_466324 | 5.57 |
ENSDART00000169691
|
mprip
|
myosin phosphatase Rho interacting protein |
| chr8_+_44783424 | 5.57 |
ENSDART00000025875
|
si:ch1073-459j12.1
|
si:ch1073-459j12.1 |
| chr25_+_19734038 | 5.51 |
ENSDART00000067354
|
zgc:101783
|
zgc:101783 |
| chr16_+_16968682 | 5.45 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr15_+_20239141 | 5.42 |
ENSDART00000101152
ENSDART00000152473 |
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr9_+_24065855 | 5.37 |
ENSDART00000161468
ENSDART00000171577 ENSDART00000172743 ENSDART00000159324 ENSDART00000079689 ENSDART00000023196 ENSDART00000101577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr5_-_42059869 | 5.36 |
ENSDART00000193984
|
cenpv
|
centromere protein V |
| chr1_-_52461322 | 5.35 |
ENSDART00000083836
|
si:ch211-217k17.7
|
si:ch211-217k17.7 |
| chr13_-_280827 | 5.28 |
ENSDART00000144819
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr22_-_5323482 | 5.25 |
ENSDART00000145785
|
s1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr17_-_51651631 | 5.24 |
ENSDART00000154699
|
ccr6b
|
chemokine (C-C motif) receptor 6b |
| chr6_-_1187565 | 5.23 |
ENSDART00000191756
|
txnrd3
|
thioredoxin reductase 3 |
| chr3_+_1015867 | 5.21 |
ENSDART00000109912
|
si:ch1073-464p5.5
|
si:ch1073-464p5.5 |
| chr25_-_13050959 | 5.19 |
ENSDART00000169041
|
ccl35.1
|
chemokine (C-C motif) ligand 35, duplicate 1 |
| chr1_-_43727418 | 5.18 |
ENSDART00000133715
ENSDART00000074597 ENSDART00000132542 ENSDART00000181792 |
bdh2
SLC9B2
|
3-hydroxybutyrate dehydrogenase, type 2 si:dkey-162b23.4 |
| chr23_+_45906137 | 5.07 |
ENSDART00000159939
|
abcg2a
|
ATP-binding cassette, sub-family G (WHITE), member 2a |
| chr12_-_18898413 | 5.06 |
ENSDART00000181281
ENSDART00000121866 |
desi1b
|
desumoylating isopeptidase 1b |
| chr2_+_25840463 | 5.04 |
ENSDART00000125178
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr10_+_8437930 | 5.03 |
ENSDART00000074553
|
pptc7b
|
PTC7 protein phosphatase homolog b |
| chr24_+_36317544 | 5.02 |
ENSDART00000048640
ENSDART00000156096 |
pus3
|
pseudouridylate synthase 3 |
| chr2_-_51512294 | 5.01 |
ENSDART00000185287
|
si:ch211-9d9.8
|
si:ch211-9d9.8 |
| chr2_+_58877162 | 4.97 |
ENSDART00000122174
|
CABZ01085658.1
|
|
| chr19_-_31035325 | 4.96 |
ENSDART00000147504
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr14_-_15739969 | 4.95 |
ENSDART00000164841
ENSDART00000192635 |
neurl1b
|
neuralized E3 ubiquitin protein ligase 1B |
| chr19_-_34117056 | 4.94 |
ENSDART00000158677
|
si:dkey-184p18.2
|
si:dkey-184p18.2 |
| chr6_-_57476465 | 4.93 |
ENSDART00000128065
|
ddx27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr5_-_16475374 | 4.92 |
ENSDART00000134274
ENSDART00000136004 |
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr2_+_25839940 | 4.85 |
ENSDART00000139927
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr1_-_9195629 | 4.84 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr12_+_13091842 | 4.81 |
ENSDART00000185477
ENSDART00000181435 ENSDART00000124799 |
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr14_+_16151636 | 4.81 |
ENSDART00000159352
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr6_-_1187749 | 4.81 |
ENSDART00000172544
|
txnrd3
|
thioredoxin reductase 3 |
| chr6_-_21189295 | 4.79 |
ENSDART00000137136
|
obsl1a
|
obscurin-like 1a |
| chr11_+_45092866 | 4.76 |
ENSDART00000163408
|
si:dkey-93h22.8
|
si:dkey-93h22.8 |
| chr16_-_38333976 | 4.74 |
ENSDART00000031895
|
cdc42se1
|
CDC42 small effector 1 |
| chr23_+_17417539 | 4.72 |
ENSDART00000182605
|
BX649300.2
|
|
| chr3_-_1387292 | 4.72 |
ENSDART00000163535
|
ddx47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr9_+_23770666 | 4.72 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr16_-_5844881 | 4.68 |
ENSDART00000085678
|
trak1a
|
trafficking protein, kinesin binding 1a |
| chr16_+_25259313 | 4.66 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr19_-_24555935 | 4.64 |
ENSDART00000132660
ENSDART00000162801 |
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr18_+_44795711 | 4.64 |
ENSDART00000110229
ENSDART00000188262 ENSDART00000139526 |
fam118b
|
family with sequence similarity 118, member B |
| chr19_+_20201254 | 4.62 |
ENSDART00000010140
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr5_-_33959868 | 4.62 |
ENSDART00000143652
|
zgc:63972
|
zgc:63972 |
| chr13_+_29238850 | 4.61 |
ENSDART00000026000
|
myofl
|
myoferlin like |
| chr15_-_36533322 | 4.59 |
ENSDART00000156466
ENSDART00000121755 |
si:dkey-262k9.4
|
si:dkey-262k9.4 |
| chr14_+_30774032 | 4.58 |
ENSDART00000139552
|
atl3
|
atlastin 3 |
| chr7_+_32021982 | 4.55 |
ENSDART00000173848
|
mettl15
|
methyltransferase like 15 |
| chr9_-_31108285 | 4.52 |
ENSDART00000003193
|
gpr183a
|
G protein-coupled receptor 183a |
| chr25_-_16589461 | 4.52 |
ENSDART00000064204
|
cpa4
|
carboxypeptidase A4 |
| chr24_-_38644937 | 4.48 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr5_-_8817458 | 4.48 |
ENSDART00000191098
|
fgf10b
|
fibroblast growth factor 10b |
| chr22_-_23591340 | 4.43 |
ENSDART00000167024
|
f13b
|
coagulation factor XIII, B polypeptide |
| chr14_+_30774515 | 4.43 |
ENSDART00000191666
|
atl3
|
atlastin 3 |
| chr17_+_38566717 | 4.43 |
ENSDART00000145147
|
sptb
|
spectrin, beta, erythrocytic |
| chr9_+_29985010 | 4.42 |
ENSDART00000020743
|
cmss1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr7_-_29164818 | 4.37 |
ENSDART00000052348
|
exosc6
|
exosome component 6 |
| chr10_-_373575 | 4.31 |
ENSDART00000114487
|
DMPK
|
DM1 protein kinase |
| chr11_+_77526 | 4.30 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr5_+_27488975 | 4.29 |
ENSDART00000123635
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr19_-_24555623 | 4.29 |
ENSDART00000176022
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr17_-_45370200 | 4.26 |
ENSDART00000186208
|
znf106a
|
zinc finger protein 106a |
| chr3_-_34027178 | 4.25 |
ENSDART00000170201
ENSDART00000151408 |
ighv1-4
ighv14-1
|
immunoglobulin heavy variable 1-4 immunoglobulin heavy variable 14-1 |
| chr18_+_38191346 | 4.22 |
ENSDART00000052703
|
nucb2b
|
nucleobindin 2b |
| chr20_+_33875256 | 4.22 |
ENSDART00000002554
|
rxrgb
|
retinoid X receptor, gamma b |
| chr23_+_26017227 | 4.21 |
ENSDART00000002939
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr5_-_37959874 | 4.17 |
ENSDART00000031719
|
mpzl2b
|
myelin protein zero-like 2b |
| chr25_+_16356083 | 4.15 |
ENSDART00000125925
ENSDART00000125444 |
tead1a
|
TEA domain family member 1a |
| chr2_-_30734098 | 4.12 |
ENSDART00000133769
|
rp1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chr5_-_29534748 | 4.07 |
ENSDART00000159587
|
AL831768.1
|
|
| chr17_+_24821627 | 4.05 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr10_+_29850330 | 4.02 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr7_+_1505507 | 4.01 |
ENSDART00000161015
|
nop10
|
NOP10 ribonucleoprotein homolog (yeast) |
| chr9_-_23944470 | 4.00 |
ENSDART00000138754
|
col6a3
|
collagen, type VI, alpha 3 |
| chr1_+_10297027 | 3.99 |
ENSDART00000152562
|
eif3f
|
eukaryotic translation initiation factor 3, subunit F |
| chr21_+_45685757 | 3.99 |
ENSDART00000160530
|
sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr16_+_33902006 | 3.98 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr8_+_54135642 | 3.97 |
ENSDART00000170712
|
brpf1
|
bromodomain and PHD finger containing, 1 |
| chr2_+_5793908 | 3.95 |
ENSDART00000145219
|
slc35d1b
|
solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1b |
| chr25_-_14424406 | 3.95 |
ENSDART00000073609
|
prmt7
|
protein arginine methyltransferase 7 |
| chr16_+_40954481 | 3.95 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr13_-_24260609 | 3.95 |
ENSDART00000138747
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr22_+_21317597 | 3.95 |
ENSDART00000132605
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr10_-_57270 | 3.94 |
ENSDART00000058411
|
hlcs
|
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase) |
| chr16_-_14074594 | 3.93 |
ENSDART00000090234
|
trim109
|
tripartite motif containing 109 |
| chr5_+_26686639 | 3.92 |
ENSDART00000079064
|
tango2
|
transport and golgi organization 2 homolog (Drosophila) |
| chr16_-_12787029 | 3.92 |
ENSDART00000139916
|
foxj2
|
forkhead box J2 |
| chr5_+_37744625 | 3.90 |
ENSDART00000014031
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr1_-_26675969 | 3.88 |
ENSDART00000054184
|
trmo
|
tRNA methyltransferase O |
Network of associatons between targets according to the STRING database.
First level regulatory network of clocka+clockb+npas2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 34.9 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 4.9 | 14.7 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 4.1 | 16.3 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 2.9 | 8.6 | GO:0098543 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 2.8 | 8.5 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 2.7 | 18.9 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 2.6 | 10.3 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 2.4 | 7.1 | GO:0042256 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 1.9 | 7.6 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 1.7 | 8.4 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 1.6 | 10.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 1.5 | 13.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 1.5 | 7.6 | GO:0043420 | anthranilate metabolic process(GO:0043420) |
| 1.5 | 15.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 1.5 | 4.5 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 1.5 | 4.4 | GO:0034475 | U4 snRNA 3'-end processing(GO:0034475) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 1.4 | 5.7 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 1.4 | 13.8 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 1.4 | 1.4 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 1.3 | 7.9 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 1.3 | 7.8 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 1.3 | 5.0 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 1.2 | 3.6 | GO:1905133 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 1.1 | 6.8 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 1.1 | 6.8 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 1.1 | 4.5 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 1.1 | 6.7 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 1.0 | 13.6 | GO:0009303 | rRNA transcription(GO:0009303) |
| 1.0 | 9.1 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 1.0 | 4.0 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 1.0 | 5.0 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 1.0 | 4.8 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 1.0 | 9.6 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.9 | 4.6 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.9 | 31.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.9 | 11.7 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.9 | 8.8 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.9 | 5.2 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.9 | 3.4 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.9 | 6.0 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.8 | 3.4 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.8 | 0.8 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.8 | 4.7 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.7 | 11.0 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.7 | 2.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.7 | 2.1 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.7 | 7.6 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.7 | 15.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.7 | 22.8 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.7 | 1.3 | GO:0034250 | positive regulation of cellular amide metabolic process(GO:0034250) |
| 0.7 | 3.3 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.6 | 1.9 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.6 | 2.5 | GO:0060585 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.6 | 1.8 | GO:0071312 | response to cocaine(GO:0042220) cellular response to alkaloid(GO:0071312) cellular response to cocaine(GO:0071314) |
| 0.6 | 1.8 | GO:0006585 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.6 | 2.4 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.6 | 2.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.6 | 8.9 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.6 | 11.8 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.6 | 2.8 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.6 | 3.3 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.5 | 12.0 | GO:0009749 | response to glucose(GO:0009749) |
| 0.5 | 8.1 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.5 | 5.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.5 | 4.7 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.5 | 1.5 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.5 | 2.6 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.5 | 8.0 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.5 | 3.5 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.5 | 6.0 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.5 | 12.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.5 | 1.9 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.5 | 10.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.5 | 3.7 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.4 | 2.6 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.4 | 2.6 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.4 | 5.9 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.4 | 33.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.4 | 23.4 | GO:0006414 | translational elongation(GO:0006414) |
| 0.4 | 71.5 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.4 | 2.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.4 | 7.1 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.4 | 1.6 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.4 | 3.7 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.4 | 2.9 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.4 | 1.1 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.3 | 1.4 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.3 | 1.3 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.3 | 3.6 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.3 | 1.3 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.3 | 2.3 | GO:0045109 | intermediate filament organization(GO:0045109) intermediate filament bundle assembly(GO:0045110) |
| 0.3 | 5.9 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.3 | 9.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.3 | 2.5 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.3 | 1.2 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.3 | 12.8 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.3 | 4.2 | GO:0032094 | response to food(GO:0032094) |
| 0.3 | 10.6 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.3 | 1.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.3 | 2.0 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.3 | 4.0 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.3 | 0.8 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.3 | 1.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.3 | 4.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.3 | 8.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.3 | 4.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 3.2 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.2 | 1.5 | GO:1902914 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 1.0 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 22.0 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.2 | 1.1 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.2 | 4.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.2 | 3.8 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.2 | 1.0 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.2 | 1.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.2 | 13.3 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.2 | 9.0 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.2 | 0.9 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.2 | 0.9 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.2 | 2.0 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.2 | 2.7 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 8.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.2 | 1.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.2 | 7.2 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.2 | 1.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 0.7 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.2 | 58.9 | GO:0006412 | translation(GO:0006412) peptide biosynthetic process(GO:0043043) |
| 0.2 | 1.7 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.2 | 18.1 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.2 | 5.9 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.2 | 4.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.2 | 2.3 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.2 | 3.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 0.9 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 3.1 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.1 | 6.9 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.1 | 7.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 6.7 | GO:0035272 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.1 | 1.9 | GO:0045762 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.1 | 4.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 2.7 | GO:0072599 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.1 | 0.4 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.1 | 0.8 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 4.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 1.4 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.1 | 1.5 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.1 | 0.7 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 2.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.8 | GO:0070375 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.1 | 8.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 0.6 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 2.3 | GO:0036294 | cellular response to decreased oxygen levels(GO:0036294) cellular response to hypoxia(GO:0071456) |
| 0.1 | 0.4 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.1 | 1.7 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.7 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.8 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 4.3 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 2.9 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 4.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 2.2 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 3.5 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 2.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 1.0 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 10.0 | GO:0006457 | protein folding(GO:0006457) |
| 0.1 | 8.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 1.9 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 4.2 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 8.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 2.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 3.6 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.1 | 3.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 1.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 2.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 2.0 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 14.5 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.1 | 3.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 4.1 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 5.3 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 1.2 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 0.3 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.1 | 0.7 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 1.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 3.2 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 10.5 | GO:0045087 | innate immune response(GO:0045087) |
| 0.1 | 2.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 1.6 | GO:0007632 | visual behavior(GO:0007632) |
| 0.1 | 0.4 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 3.9 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.1 | 1.2 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.1 | 2.1 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 19.4 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.1 | 1.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.7 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 0.5 | GO:0030317 | sperm motility(GO:0030317) |
| 0.1 | 3.2 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 4.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 2.5 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 3.6 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.1 | 2.8 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.4 | GO:0021707 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 1.0 | GO:0031577 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 3.1 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 1.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 1.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.9 | GO:0032508 | DNA geometric change(GO:0032392) DNA duplex unwinding(GO:0032508) |
| 0.0 | 5.7 | GO:0043066 | negative regulation of apoptotic process(GO:0043066) |
| 0.0 | 0.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 1.3 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 1.0 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 1.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 3.8 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 1.1 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 1.6 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 1.1 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 2.2 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.0 | 0.5 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 6.4 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 1.1 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.0 | 1.9 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 1.3 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.7 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 1.2 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 1.4 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 2.9 | GO:0042981 | regulation of apoptotic process(GO:0042981) |
| 0.0 | 0.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.3 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.3 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 11.3 | GO:0006508 | proteolysis(GO:0006508) |
| 0.0 | 0.4 | GO:0036293 | response to hypoxia(GO:0001666) response to decreased oxygen levels(GO:0036293) response to oxygen levels(GO:0070482) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 14.7 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 3.5 | 10.6 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 2.9 | 11.7 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 2.2 | 34.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 1.8 | 22.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 1.8 | 8.9 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 1.7 | 5.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 1.6 | 4.8 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 1.4 | 5.7 | GO:0097268 | cytoophidium(GO:0097268) |
| 1.2 | 3.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 1.1 | 4.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 1.0 | 64.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 1.0 | 4.0 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 1.0 | 4.9 | GO:1990923 | PET complex(GO:1990923) |
| 0.9 | 10.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.9 | 3.7 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.9 | 2.7 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.9 | 4.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.9 | 10.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.8 | 2.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.8 | 2.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.7 | 11.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.7 | 6.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.7 | 3.3 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.6 | 4.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.6 | 9.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.5 | 3.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.5 | 8.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.4 | 6.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.4 | 3.3 | GO:0030428 | cell septum(GO:0030428) |
| 0.4 | 4.4 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.4 | 7.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.4 | 5.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.4 | 1.9 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.4 | 96.7 | GO:0005730 | nucleolus(GO:0005730) |
| 0.3 | 13.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.3 | 20.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.3 | 3.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.3 | 2.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.3 | 1.5 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.3 | 1.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.3 | 1.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.3 | 1.8 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 1.0 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.2 | 4.0 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 1.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 1.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 0.9 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 2.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 1.8 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.2 | 9.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 1.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 2.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 4.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 3.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.2 | 1.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.2 | 1.0 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 1.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 0.8 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 15.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 1.8 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 7.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 2.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 5.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 17.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 6.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 2.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.7 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 5.0 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 2.5 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 2.7 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.1 | 1.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 9.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 29.3 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.1 | 1.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 4.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 6.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 6.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 1.3 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 1.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.8 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 1.9 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 3.5 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.2 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 2.1 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 28.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 33.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.4 | GO:0032421 | stereocilium bundle(GO:0032421) |
| 0.0 | 16.3 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.2 | GO:0042613 | MHC protein complex(GO:0042611) MHC class II protein complex(GO:0042613) |
| 0.0 | 1.6 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.0 | 6.8 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 15.9 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 1.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.6 | GO:0004776 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 3.4 | 13.7 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 2.9 | 11.7 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 2.9 | 8.6 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 2.8 | 8.5 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 2.6 | 10.3 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 2.4 | 9.7 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 2.4 | 7.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 2.3 | 16.3 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 2.2 | 8.8 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 1.9 | 7.6 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 1.8 | 5.4 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 1.7 | 6.8 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 1.6 | 7.8 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 1.5 | 9.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 1.4 | 5.7 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 1.3 | 8.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 1.3 | 4.0 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 1.3 | 7.8 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 1.3 | 8.9 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 1.3 | 5.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 1.2 | 7.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 1.1 | 6.8 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 1.1 | 10.0 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 1.0 | 9.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 1.0 | 4.0 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.9 | 42.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.8 | 7.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.8 | 2.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.8 | 7.6 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.8 | 4.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.7 | 5.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.7 | 13.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.7 | 5.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.6 | 28.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.6 | 2.5 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.6 | 4.9 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.6 | 1.8 | GO:0016496 | substance P receptor activity(GO:0016496) |
| 0.6 | 1.8 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.6 | 11.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.6 | 1.7 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.6 | 10.7 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.6 | 11.8 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.5 | 1.1 | GO:1901474 | drug transmembrane transporter activity(GO:0015238) azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.5 | 22.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.5 | 2.6 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.5 | 7.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.5 | 8.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.5 | 5.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.5 | 7.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.5 | 2.8 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.4 | 1.3 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.4 | 3.1 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.4 | 3.5 | GO:0005113 | patched binding(GO:0005113) |
| 0.4 | 1.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.4 | 14.5 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.4 | 2.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.4 | 3.3 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.4 | 10.9 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.4 | 2.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.4 | 4.5 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.4 | 4.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.4 | 6.0 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.4 | 28.7 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.3 | 1.4 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.3 | 1.4 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.3 | 1.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.3 | 1.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 1.8 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.3 | 2.4 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.3 | 7.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.3 | 8.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 4.2 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.3 | 2.7 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.3 | 5.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.3 | 48.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.3 | 9.1 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.3 | 2.9 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.3 | 4.0 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.3 | 2.0 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.3 | 3.6 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.3 | 1.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.3 | 2.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.3 | 6.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.3 | 1.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 27.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 2.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.2 | 0.9 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.2 | 0.7 | GO:0034618 | arginine binding(GO:0034618) |
| 0.2 | 2.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 2.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 1.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.2 | 1.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 7.3 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.2 | 5.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.2 | 8.2 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 27.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 7.0 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 3.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 1.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 1.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 0.8 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.2 | 1.9 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.7 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 1.0 | GO:0016918 | retinal binding(GO:0016918) retinol transporter activity(GO:0034632) |
| 0.1 | 3.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.8 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.1 | 13.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 6.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 2.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 1.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 9.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 3.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 3.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 2.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 7.6 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 4.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 2.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.5 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.4 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 1.2 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 13.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 2.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 4.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 3.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 2.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 3.4 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.1 | 1.0 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 5.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 2.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.3 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 1.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 1.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 2.2 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.1 | 2.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 11.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 1.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 2.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 15.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.7 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 5.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.1 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.6 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 2.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.4 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 3.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 16.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 3.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 7.8 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.6 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 2.7 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 1.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 1.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.8 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 1.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 8.3 | GO:0008236 | serine-type peptidase activity(GO:0008236) serine hydrolase activity(GO:0017171) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 20.0 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.5 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 2.5 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 1.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 4.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 4.7 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 1.9 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 4.1 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.0 | 7.7 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.6 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.5 | 5.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.4 | 6.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.4 | 8.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.4 | 6.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.4 | 11.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.3 | 5.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 3.9 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.2 | 3.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 7.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 3.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 9.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 6.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 11.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 4.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.2 | 7.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.2 | 8.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 3.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 10.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 5.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 2.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 1.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.8 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 5.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 23.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 1.4 | 22.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.7 | 21.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.7 | 7.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.7 | 7.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.6 | 71.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.6 | 6.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.6 | 9.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.6 | 7.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.6 | 7.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.6 | 10.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.5 | 11.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.5 | 4.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 8.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.4 | 11.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.4 | 8.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.3 | 8.3 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.3 | 4.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.3 | 11.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.3 | 4.4 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.3 | 3.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.3 | 4.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 5.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 2.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 6.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.2 | 2.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 1.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 1.9 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 2.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 3.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 3.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 1.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 8.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 3.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 11.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 3.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 1.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 2.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.1 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.1 | 4.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 2.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 4.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.0 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 0.8 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.1 | 4.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.0 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.1 | 1.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 3.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 5.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 0.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 2.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.0 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 2.3 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 2.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 2.1 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 1.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |