Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
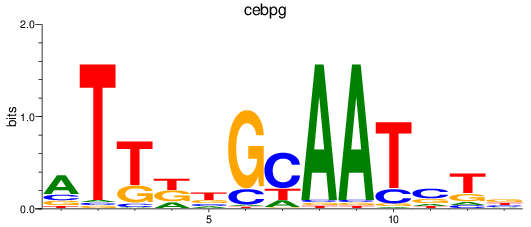
Results for cebpg
Z-value: 1.58
Transcription factors associated with cebpg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cebpg
|
ENSDARG00000036073 | CCAAT enhancer binding protein gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cebpg | dr11_v1_chr7_+_38090515_38090515 | 0.16 | 1.3e-01 | Click! |
Activity profile of cebpg motif
Sorted Z-values of cebpg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_10071422 | 49.43 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr22_-_23668356 | 43.81 |
ENSDART00000167106
ENSDART00000159622 ENSDART00000163228 |
cfh
|
complement factor H |
| chr5_-_63509581 | 33.58 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr20_-_40755614 | 32.21 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr22_-_26175237 | 30.77 |
ENSDART00000108737
|
c3b.2
|
complement component c3b, tandem duplicate 2 |
| chr8_-_50147948 | 29.77 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr5_+_45677781 | 26.40 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr22_-_26236188 | 26.29 |
ENSDART00000162640
ENSDART00000167169 ENSDART00000138595 |
c3b.1
|
complement component c3b, tandem duplicate 1 |
| chr12_-_22524388 | 21.36 |
ENSDART00000020942
|
shbg
|
sex hormone-binding globulin |
| chr22_-_24757785 | 21.08 |
ENSDART00000078225
|
vtg5
|
vitellogenin 5 |
| chr24_-_33284945 | 20.49 |
ENSDART00000155429
ENSDART00000112845 |
zgc:195173
|
zgc:195173 |
| chr6_-_609880 | 20.11 |
ENSDART00000149248
ENSDART00000148867 ENSDART00000149414 ENSDART00000148552 ENSDART00000148391 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr20_+_23440632 | 19.22 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr16_+_23984179 | 16.74 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr9_+_38292947 | 14.86 |
ENSDART00000146663
|
tfcp2l1
|
transcription factor CP2-like 1 |
| chr15_-_29348212 | 14.45 |
ENSDART00000133117
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr19_-_34979837 | 13.84 |
ENSDART00000044838
|
ndrg1a
|
N-myc downstream regulated 1a |
| chr23_-_31763753 | 13.31 |
ENSDART00000053399
|
aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr8_-_51293265 | 12.83 |
ENSDART00000127875
ENSDART00000181145 |
bmp1a
|
bone morphogenetic protein 1a |
| chr10_-_22803740 | 12.69 |
ENSDART00000079469
ENSDART00000187968 ENSDART00000122543 |
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr1_+_14253118 | 12.31 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr7_+_56577906 | 11.59 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr16_+_48753664 | 11.18 |
ENSDART00000155148
|
si:ch73-31d8.2
|
si:ch73-31d8.2 |
| chr7_-_58289847 | 11.09 |
ENSDART00000012822
|
CR354540.1
|
|
| chr17_-_10838434 | 10.85 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr15_-_21014270 | 10.63 |
ENSDART00000154019
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr12_+_46386983 | 10.52 |
ENSDART00000183982
|
BX005305.3
|
Danio rerio legumain (LOC100005356), mRNA. |
| chr1_+_17892944 | 10.37 |
ENSDART00000013021
|
tlr3
|
toll-like receptor 3 |
| chr14_-_11507211 | 9.96 |
ENSDART00000186873
ENSDART00000109181 ENSDART00000186166 ENSDART00000186986 |
zgc:174917
|
zgc:174917 |
| chr2_+_4207209 | 9.86 |
ENSDART00000157903
ENSDART00000166476 |
gata6
|
GATA binding protein 6 |
| chr4_-_9549693 | 9.85 |
ENSDART00000160242
|
FQ377934.1
|
|
| chr12_+_25775734 | 9.85 |
ENSDART00000024415
ENSDART00000149198 |
epas1a
|
endothelial PAS domain protein 1a |
| chr12_+_46404307 | 9.80 |
ENSDART00000185011
|
BX005305.4
|
|
| chr21_+_17051478 | 9.55 |
ENSDART00000047201
ENSDART00000161650 ENSDART00000167298 |
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr1_+_31674297 | 9.17 |
ENSDART00000044214
|
wbp1lb
|
WW domain binding protein 1-like b |
| chr8_-_53488832 | 8.92 |
ENSDART00000191801
|
chdh
|
choline dehydrogenase |
| chr12_-_33770299 | 8.84 |
ENSDART00000189849
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr19_-_15192638 | 8.83 |
ENSDART00000048151
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr9_+_426392 | 8.76 |
ENSDART00000172515
|
bzw1b
|
basic leucine zipper and W2 domains 1b |
| chr5_-_69944084 | 8.56 |
ENSDART00000188557
ENSDART00000127782 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr10_-_7785930 | 8.52 |
ENSDART00000043961
ENSDART00000111058 |
mpx
|
myeloid-specific peroxidase |
| chr12_+_46462090 | 8.06 |
ENSDART00000130748
|
BX005305.2
|
|
| chr15_-_23793641 | 7.87 |
ENSDART00000122891
|
tmem97
|
transmembrane protein 97 |
| chr10_-_36808348 | 7.76 |
ENSDART00000099320
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr20_-_5369105 | 7.55 |
ENSDART00000114316
|
sptlc2b
|
serine palmitoyltransferase, long chain base subunit 2b |
| chr24_+_38671054 | 7.40 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr25_-_29415369 | 7.25 |
ENSDART00000110774
ENSDART00000019183 |
ugt5a2
ugt5a1
|
UDP glucuronosyltransferase 5 family, polypeptide A2 UDP glucuronosyltransferase 5 family, polypeptide A1 |
| chr12_+_46483618 | 6.92 |
ENSDART00000186970
|
BX005305.5
|
|
| chr6_+_10333920 | 6.77 |
ENSDART00000151667
ENSDART00000151477 |
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr12_+_46443477 | 6.68 |
ENSDART00000191873
|
BX005305.1
|
|
| chr15_-_3282220 | 6.64 |
ENSDART00000092942
|
slc25a15a
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15a |
| chr12_+_46425800 | 6.46 |
ENSDART00000191965
|
BX005305.1
|
|
| chr14_-_45551572 | 6.29 |
ENSDART00000111410
|
ganab
|
glucosidase, alpha; neutral AB |
| chr12_+_46512881 | 6.25 |
ENSDART00000105454
|
CU855711.1
|
|
| chr2_-_40199780 | 6.11 |
ENSDART00000113901
|
ccl34a.4
|
chemokine (C-C motif) ligand 34a, duplicate 4 |
| chr14_-_38929885 | 6.05 |
ENSDART00000148737
|
btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chr23_+_21405201 | 5.98 |
ENSDART00000144409
|
iffo2a
|
intermediate filament family orphan 2a |
| chr2_-_30182353 | 5.89 |
ENSDART00000019149
|
rpl7
|
ribosomal protein L7 |
| chr14_-_43602968 | 5.87 |
ENSDART00000108825
|
adh5
|
alcohol dehydrogenase 5 |
| chr10_-_28193642 | 5.74 |
ENSDART00000019050
|
rps6kb1a
|
ribosomal protein S6 kinase b, polypeptide 1a |
| chr5_+_4298636 | 5.73 |
ENSDART00000100061
|
prdx4
|
peroxiredoxin 4 |
| chr9_-_21838045 | 5.68 |
ENSDART00000147471
|
acod1
|
aconitate decarboxylase 1 |
| chr10_-_41980797 | 5.63 |
ENSDART00000076575
|
rhof
|
ras homolog family member F |
| chr9_-_53062083 | 5.51 |
ENSDART00000122155
|
zmp:0000000936
|
zmp:0000000936 |
| chr1_+_58067815 | 5.37 |
ENSDART00000156678
|
si:ch211-114l13.4
|
si:ch211-114l13.4 |
| chr6_-_39344259 | 5.37 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr13_-_30996072 | 5.24 |
ENSDART00000181661
|
wdfy4
|
WDFY family member 4 |
| chr24_-_38644937 | 5.16 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr1_+_45056371 | 5.00 |
ENSDART00000073689
ENSDART00000167309 |
btr01
|
bloodthirsty-related gene family, member 1 |
| chr16_-_42004544 | 4.82 |
ENSDART00000034544
|
caspa
|
caspase a |
| chr8_-_2602572 | 4.63 |
ENSDART00000110482
|
zdhhc12a
|
zinc finger, DHHC-type containing 12a |
| chr7_+_33152723 | 4.62 |
ENSDART00000132658
|
si:ch211-194p6.10
|
si:ch211-194p6.10 |
| chr6_-_39649504 | 4.61 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr5_-_23783739 | 4.56 |
ENSDART00000139502
|
GBGT1 (1 of many)
|
si:ch211-287c22.1 |
| chr24_-_32665283 | 4.54 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr23_+_27675581 | 4.52 |
ENSDART00000127198
|
rps26
|
ribosomal protein S26 |
| chr25_+_32474031 | 4.46 |
ENSDART00000152124
|
sqor
|
sulfide quinone oxidoreductase |
| chr14_-_38865800 | 4.34 |
ENSDART00000173047
|
gsr
|
glutathione reductase |
| chr9_-_34885902 | 4.33 |
ENSDART00000137504
|
p2ry8
|
P2Y receptor family member 8 |
| chr18_+_13300768 | 4.32 |
ENSDART00000137506
|
si:ch211-260p9.3
|
si:ch211-260p9.3 |
| chr4_-_5595237 | 4.26 |
ENSDART00000109854
|
vegfab
|
vascular endothelial growth factor Ab |
| chr21_-_16219400 | 4.13 |
ENSDART00000124871
|
tnfrsfa
|
tumor necrosis factor receptor superfamily, member a |
| chr19_+_7045033 | 4.10 |
ENSDART00000146579
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr19_+_16015881 | 4.05 |
ENSDART00000187135
|
ctps1a
|
CTP synthase 1a |
| chr9_+_9502610 | 4.03 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr1_+_44800298 | 3.94 |
ENSDART00000073712
ENSDART00000187054 |
tmem187
|
transmembrane protein 187 |
| chr12_-_16990896 | 3.86 |
ENSDART00000152402
|
ifit12
|
interferon-induced protein with tetratricopeptide repeats 12 |
| chr1_+_55752593 | 3.86 |
ENSDART00000108838
ENSDART00000134770 |
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr3_-_38745598 | 3.85 |
ENSDART00000127483
|
si:ch211-198m17.1
|
si:ch211-198m17.1 |
| chr17_+_51744450 | 3.83 |
ENSDART00000190955
ENSDART00000149807 |
odc1
|
ornithine decarboxylase 1 |
| chr21_-_11646878 | 3.73 |
ENSDART00000162426
ENSDART00000135937 ENSDART00000131448 ENSDART00000148097 ENSDART00000133443 |
cast
|
calpastatin |
| chr11_+_43419809 | 3.73 |
ENSDART00000172982
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr10_-_23099809 | 3.67 |
ENSDART00000148333
ENSDART00000079703 ENSDART00000162444 |
nle1
|
notchless homolog 1 (Drosophila) |
| chr7_-_16596938 | 3.67 |
ENSDART00000134548
|
e2f8
|
E2F transcription factor 8 |
| chr8_-_4475908 | 3.57 |
ENSDART00000191027
|
si:ch211-166a6.5
|
si:ch211-166a6.5 |
| chr13_-_24260609 | 3.57 |
ENSDART00000138747
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr21_+_43178831 | 3.51 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr11_-_31172276 | 3.48 |
ENSDART00000171520
|
si:dkey-238i5.3
|
si:dkey-238i5.3 |
| chr20_-_30900947 | 3.47 |
ENSDART00000153419
ENSDART00000062536 |
hebp2
|
heme binding protein 2 |
| chr24_-_26485098 | 3.42 |
ENSDART00000135496
ENSDART00000009609 ENSDART00000133782 ENSDART00000141029 ENSDART00000113739 |
eif5a
|
eukaryotic translation initiation factor 5A |
| chr7_+_49862837 | 3.37 |
ENSDART00000174315
|
slc1a2a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2a |
| chr17_+_51743908 | 3.35 |
ENSDART00000149039
ENSDART00000148869 |
odc1
|
ornithine decarboxylase 1 |
| chr15_+_38018301 | 3.33 |
ENSDART00000114188
ENSDART00000171150 |
si:ch73-380l3.1
|
si:ch73-380l3.1 |
| chr19_-_20403318 | 3.32 |
ENSDART00000136826
|
dazl
|
deleted in azoospermia-like |
| chr3_+_54581987 | 3.32 |
ENSDART00000018071
|
eif3g
|
eukaryotic translation initiation factor 3, subunit G |
| chr7_-_16597130 | 3.28 |
ENSDART00000144118
|
e2f8
|
E2F transcription factor 8 |
| chr3_-_31254979 | 3.13 |
ENSDART00000130280
|
apnl
|
actinoporin-like protein |
| chr2_+_25378457 | 3.12 |
ENSDART00000089108
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr10_+_23099890 | 3.12 |
ENSDART00000135890
|
LTN1
|
si:dkey-175g6.5 |
| chr5_-_24238733 | 3.04 |
ENSDART00000138170
|
plscr3a
|
phospholipid scramblase 3a |
| chr13_-_24257631 | 2.96 |
ENSDART00000146524
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr5_+_54400971 | 2.96 |
ENSDART00000169695
|
bspry
|
B-box and SPRY domain containing |
| chr25_-_16076257 | 2.96 |
ENSDART00000140780
|
ovch2
|
ovochymase 2 |
| chr2_+_37971353 | 2.84 |
ENSDART00000165085
|
apol1
|
apolipoprotein L, 1 |
| chr18_-_25276932 | 2.67 |
ENSDART00000076183
|
plin1
|
perilipin 1 |
| chr4_+_9508505 | 2.66 |
ENSDART00000080842
|
kitlgb
|
kit ligand b |
| chr9_-_9419704 | 2.62 |
ENSDART00000138996
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr13_-_14929236 | 2.62 |
ENSDART00000020576
|
cdc25b
|
cell division cycle 25B |
| chr19_-_2822372 | 2.61 |
ENSDART00000109130
ENSDART00000122385 |
recql4
|
RecQ helicase-like 4 |
| chr20_+_29436601 | 2.59 |
ENSDART00000136804
|
fmn1
|
formin 1 |
| chr4_+_17279966 | 2.56 |
ENSDART00000067005
ENSDART00000137487 |
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr1_+_30551777 | 2.50 |
ENSDART00000112778
|
gpr183b
|
G protein-coupled receptor 183b |
| chr15_+_7064819 | 2.48 |
ENSDART00000155268
|
pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr3_+_43086548 | 2.47 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr18_-_26797723 | 2.45 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr19_+_16016038 | 2.40 |
ENSDART00000131319
|
ctps1a
|
CTP synthase 1a |
| chr19_-_10915898 | 2.32 |
ENSDART00000163179
|
pip5k1aa
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, a |
| chr3_-_23574622 | 2.31 |
ENSDART00000176012
|
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr11_-_34783938 | 2.21 |
ENSDART00000135725
ENSDART00000039847 |
chchd4a
|
coiled-coil-helix-coiled-coil-helix domain containing 4a |
| chr20_+_25879826 | 2.20 |
ENSDART00000018519
|
zgc:153896
|
zgc:153896 |
| chr8_-_36140405 | 2.17 |
ENSDART00000182806
|
CT583723.2
|
|
| chr2_+_42247560 | 2.17 |
ENSDART00000101230
ENSDART00000143094 |
ftr06
|
finTRIM family, member 6 |
| chr17_-_23603263 | 2.10 |
ENSDART00000079559
|
ifit16
|
interferon-induced protein with tetratricopeptide repeats 16 |
| chr2_+_11029138 | 2.09 |
ENSDART00000138737
ENSDART00000081058 ENSDART00000153662 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr17_+_28670132 | 2.08 |
ENSDART00000076344
ENSDART00000164981 ENSDART00000182851 |
hectd1
|
HECT domain containing 1 |
| chr4_-_20313810 | 2.07 |
ENSDART00000136350
|
dcp1b
|
decapping mRNA 1B |
| chr1_-_28885919 | 2.04 |
ENSDART00000152182
|
poglut1
|
protein O-glucosyltransferase 1 |
| chr8_+_22955262 | 1.99 |
ENSDART00000193806
|
zgc:136605
|
zgc:136605 |
| chr21_-_14762944 | 1.97 |
ENSDART00000114096
|
arrdc1b
|
arrestin domain containing 1b |
| chr20_+_32118559 | 1.96 |
ENSDART00000026273
|
cd164
|
CD164 molecule, sialomucin |
| chr2_-_9696283 | 1.95 |
ENSDART00000165712
|
selenot1a
|
selenoprotein T, 1a |
| chr4_-_20292821 | 1.86 |
ENSDART00000136069
ENSDART00000192504 |
cacna2d4a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4a |
| chr8_+_22955478 | 1.75 |
ENSDART00000099911
|
zgc:136605
|
zgc:136605 |
| chr1_+_2260407 | 1.73 |
ENSDART00000058876
|
kpnb3
|
karyopherin (importin) beta 3 |
| chr7_-_25934540 | 1.70 |
ENSDART00000192761
|
BX005022.2
|
|
| chr12_-_35949936 | 1.64 |
ENSDART00000192583
|
AL954682.1
|
|
| chr17_-_20218357 | 1.61 |
ENSDART00000155990
ENSDART00000155632 ENSDART00000156540 ENSDART00000063523 |
mgmt
|
O-6-methylguanine-DNA methyltransferase |
| chr4_+_42604252 | 1.61 |
ENSDART00000184850
|
CR925768.1
|
|
| chr6_+_8315050 | 1.60 |
ENSDART00000189987
|
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr21_+_26535034 | 1.49 |
ENSDART00000180709
|
vegfbb
|
vascular endothelial growth factor Bb |
| chr5_+_41510387 | 1.47 |
ENSDART00000023779
|
vcp
|
valosin containing protein |
| chr9_-_12874774 | 1.43 |
ENSDART00000131385
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr1_+_41131481 | 1.37 |
ENSDART00000145272
|
lrpap1
|
low density lipoprotein receptor-related protein associated protein 1 |
| chr3_-_34547000 | 1.26 |
ENSDART00000166623
|
sept9a
|
septin 9a |
| chr13_-_50463938 | 1.26 |
ENSDART00000083857
|
ccnj
|
cyclin J |
| chr6_+_10077756 | 1.22 |
ENSDART00000150982
|
htr2ab
|
5-hydroxytryptamine (serotonin) receptor 2A, genome duplicate b |
| chr24_+_16905188 | 1.19 |
ENSDART00000066760
|
cct5
|
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr5_+_33519943 | 1.18 |
ENSDART00000131316
|
ms4a17c.1
|
membrane-spanning 4-domains, subfamily A, member 17C.1 |
| chr3_+_34986837 | 1.16 |
ENSDART00000190341
|
smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr16_-_44945224 | 1.15 |
ENSDART00000156921
|
ncam3
|
neural cell adhesion molecule 3 |
| chr21_-_40174647 | 1.12 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr15_+_31296517 | 1.05 |
ENSDART00000132518
|
or116-1
|
odorant receptor, family F, subfamily 116, member 1 |
| chr3_-_23575007 | 1.00 |
ENSDART00000155282
ENSDART00000087726 |
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr9_+_50000504 | 0.98 |
ENSDART00000164409
ENSDART00000165451 ENSDART00000166509 |
slc38a11
|
solute carrier family 38, member 11 |
| chr25_+_6122823 | 0.98 |
ENSDART00000191824
ENSDART00000067514 |
rbpms2a
|
RNA binding protein with multiple splicing 2a |
| chr13_+_30903816 | 0.95 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr15_+_31303355 | 0.94 |
ENSDART00000143423
|
or116-2
|
odorant receptor, family F, subfamily 116, member 2 |
| chr13_+_29193649 | 0.94 |
ENSDART00000045951
|
unc5b
|
unc-5 netrin receptor B |
| chr4_-_73227710 | 0.94 |
ENSDART00000193165
|
LO018260.3
|
|
| chr18_+_32844439 | 0.93 |
ENSDART00000166038
|
olfcg1
|
olfactory receptor C family, g1 |
| chr25_+_20716176 | 0.92 |
ENSDART00000073651
|
ergic2
|
ERGIC and golgi 2 |
| chr4_-_37479395 | 0.91 |
ENSDART00000168299
ENSDART00000157864 |
si:dkey-106c17.2
|
si:dkey-106c17.2 |
| chr25_+_20715950 | 0.88 |
ENSDART00000180223
|
ergic2
|
ERGIC and golgi 2 |
| chr21_-_45077429 | 0.88 |
ENSDART00000187268
ENSDART00000191003 |
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr17_+_43908428 | 0.87 |
ENSDART00000180332
|
msh4
|
mutS homolog 4 |
| chr22_-_10487490 | 0.86 |
ENSDART00000064798
|
aspn
|
asporin (LRR class 1) |
| chr20_+_4051752 | 0.86 |
ENSDART00000092180
|
exoc8
|
exocyst complex component 8 |
| chr22_-_16317886 | 0.82 |
ENSDART00000163664
|
trmt13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr9_-_40664923 | 0.77 |
ENSDART00000135134
|
bard1
|
BRCA1 associated RING domain 1 |
| chr18_-_15551360 | 0.75 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr7_+_38089650 | 0.75 |
ENSDART00000052365
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr15_+_28355023 | 0.74 |
ENSDART00000122159
|
si:dkey-118k5.3
|
si:dkey-118k5.3 |
| chr16_+_28601974 | 0.60 |
ENSDART00000018235
|
crot
|
carnitine O-octanoyltransferase |
| chr23_-_16843649 | 0.60 |
ENSDART00000129971
|
si:dkey-63j12.4
|
si:dkey-63j12.4 |
| chr14_+_7377552 | 0.59 |
ENSDART00000142158
ENSDART00000141471 |
hars
|
histidyl-tRNA synthetase |
| chr9_-_32300611 | 0.58 |
ENSDART00000127938
|
hspd1
|
heat shock 60 protein 1 |
| chr23_-_11130683 | 0.57 |
ENSDART00000181189
|
cntn3a.2
|
contactin 3a, tandem duplicate 2 |
| chr5_+_18012154 | 0.56 |
ENSDART00000139431
ENSDART00000048859 |
ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr7_-_69121896 | 0.56 |
ENSDART00000130227
|
crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr18_+_5510251 | 0.54 |
ENSDART00000007797
|
slc30a4
|
solute carrier family 30 (zinc transporter), member 4 |
| chr24_-_21507627 | 0.50 |
ENSDART00000170984
|
rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr12_+_22657925 | 0.45 |
ENSDART00000153048
|
si:dkey-219e21.4
|
si:dkey-219e21.4 |
| chr4_+_37936249 | 0.44 |
ENSDART00000184022
|
si:dkeyp-82b4.2
|
si:dkeyp-82b4.2 |
| chr9_+_44397151 | 0.43 |
ENSDART00000185947
|
ssfa2
|
sperm specific antigen 2 |
| chr6_+_27452289 | 0.39 |
ENSDART00000186265
|
sned1
|
sushi, nidogen and EGF-like domains 1 |
| chr4_+_8670662 | 0.38 |
ENSDART00000168768
|
adipor2
|
adiponectin receptor 2 |
| chr3_+_40809011 | 0.38 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr3_-_57666518 | 0.35 |
ENSDART00000102062
|
timp2b
|
TIMP metallopeptidase inhibitor 2b |
| chr23_-_40814080 | 0.30 |
ENSDART00000135713
|
si:dkeyp-27c8.1
|
si:dkeyp-27c8.1 |
| chr7_-_13884610 | 0.29 |
ENSDART00000006897
|
rlbp1a
|
retinaldehyde binding protein 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of cebpg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.3 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 4.1 | 12.3 | GO:0002432 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 3.6 | 14.4 | GO:0003418 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 3.5 | 10.4 | GO:0032602 | chemokine production(GO:0032602) toll-like receptor 3 signaling pathway(GO:0034138) |
| 3.3 | 49.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 3.0 | 8.9 | GO:0019695 | choline metabolic process(GO:0019695) |
| 2.2 | 6.6 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 2.2 | 10.9 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 2.1 | 12.8 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 2.0 | 13.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 2.0 | 9.9 | GO:0043011 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 1.8 | 8.8 | GO:0010719 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 1.7 | 6.9 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 1.6 | 6.5 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 1.5 | 4.5 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 1.4 | 10.0 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 1.4 | 1.4 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 1.3 | 8.8 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 1.1 | 20.5 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 1.1 | 4.5 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 1.1 | 14.2 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 1.0 | 4.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.9 | 26.4 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.9 | 16.7 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.8 | 5.9 | GO:0046292 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.8 | 7.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.7 | 2.1 | GO:0070589 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.7 | 7.5 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.6 | 2.6 | GO:0009099 | leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.6 | 3.7 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.6 | 3.3 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.5 | 21.1 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.5 | 3.7 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.5 | 2.6 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.5 | 5.7 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.5 | 3.1 | GO:0051818 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.5 | 2.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.5 | 1.5 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.5 | 3.4 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.4 | 12.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.4 | 9.8 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.4 | 7.8 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.3 | 2.1 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.3 | 2.7 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.3 | 5.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.3 | 1.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.3 | 3.6 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.3 | 2.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.3 | 2.7 | GO:0010890 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.2 | 1.0 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.2 | 2.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 4.3 | GO:0010634 | positive regulation of epithelial cell migration(GO:0010634) |
| 0.2 | 3.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 6.5 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.2 | 4.8 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.1 | 6.1 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.1 | 20.7 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.1 | 2.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.9 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 4.0 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 1.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 2.6 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 2.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 3.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 4.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 3.9 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 1.2 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.1 | 1.9 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 0.5 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 4.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 2.8 | GO:0035775 | pronephric glomerulus morphogenesis(GO:0035775) |
| 0.1 | 0.9 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 1.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.9 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 1.2 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.1 | 0.6 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 6.6 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 3.9 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 6.1 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 9.5 | GO:0001947 | heart looping(GO:0001947) |
| 0.1 | 5.7 | GO:0031929 | TOR signaling(GO:0031929) |
| 0.1 | 2.1 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 2.5 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 3.0 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 3.2 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 2.0 | GO:0003014 | renal system process(GO:0003014) |
| 0.0 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.4 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 5.6 | GO:0007163 | establishment or maintenance of cell polarity(GO:0007163) |
| 0.0 | 1.7 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 3.8 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 38.8 | GO:0006508 | proteolysis(GO:0006508) |
| 0.0 | 1.3 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.6 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.8 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 1.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 1.3 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.1 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.5 | 49.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.7 | 16.7 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 1.6 | 6.5 | GO:0097268 | cytoophidium(GO:0097268) |
| 1.4 | 10.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 1.2 | 4.8 | GO:0061702 | inflammasome complex(GO:0061702) |
| 1.1 | 3.4 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 1.0 | 3.1 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.8 | 7.5 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.7 | 2.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.6 | 2.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.5 | 5.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.4 | 3.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.4 | 9.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.4 | 1.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.3 | 3.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 8.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.2 | 224.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.2 | 3.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.1 | 11.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 1.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 5.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 4.5 | GO:0022626 | cytosolic ribosome(GO:0022626) cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.8 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 1.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 2.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 3.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 6.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 2.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 32.0 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 2.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 8.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.3 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.2 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 1.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.8 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 1.2 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 12.6 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.8 | 26.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 3.1 | 31.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 3.1 | 12.3 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 3.0 | 21.1 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 1.6 | 6.5 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 1.5 | 4.5 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 1.2 | 5.9 | GO:0051903 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 1.1 | 5.5 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 1.1 | 7.5 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 1.1 | 4.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.9 | 6.9 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.9 | 4.3 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.8 | 5.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.8 | 7.2 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.8 | 7.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.7 | 6.6 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.7 | 2.7 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.6 | 2.6 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.6 | 104.6 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.6 | 2.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.6 | 9.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.5 | 8.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.5 | 5.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.5 | 8.8 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.4 | 1.6 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.4 | 2.0 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.4 | 3.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.3 | 4.8 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.3 | 6.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 21.4 | GO:0042562 | hormone binding(GO:0042562) |
| 0.3 | 13.3 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.3 | 2.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.3 | 2.0 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.3 | 2.6 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.3 | 15.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 4.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.3 | 3.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 1.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 2.5 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.2 | 1.9 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.2 | 1.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.2 | 46.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 4.6 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.2 | 3.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 10.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.2 | 4.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 1.6 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 8.5 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.2 | 5.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 1.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.1 | 10.0 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 6.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 4.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 1.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.6 | GO:0016415 | octanoyltransferase activity(GO:0016415) |
| 0.1 | 2.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 1.7 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 4.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.9 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 3.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.1 | 14.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 12.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 6.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 6.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 8.6 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.1 | 3.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 10.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.2 | GO:0034246 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.1 | 0.9 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 2.1 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 54.4 | GO:0005102 | receptor binding(GO:0005102) |
| 0.0 | 3.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 5.6 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 6.6 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.6 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.8 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 3.5 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.9 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 6.6 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 0.6 | GO:0043130 | ubiquitin binding(GO:0043130) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 49.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.6 | 21.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.5 | 10.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 6.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 9.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 2.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 2.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 2.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.2 | 9.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 15.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 2.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.0 | 43.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 4.5 | 49.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.7 | 16.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 1.5 | 33.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 1.2 | 10.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.9 | 26.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.4 | 4.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.4 | 6.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.3 | 2.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 2.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.2 | 2.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.2 | 4.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 6.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 7.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.2 | 2.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 7.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 3.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 2.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 9.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 0.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 4.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 5.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 5.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.6 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.3 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |