Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
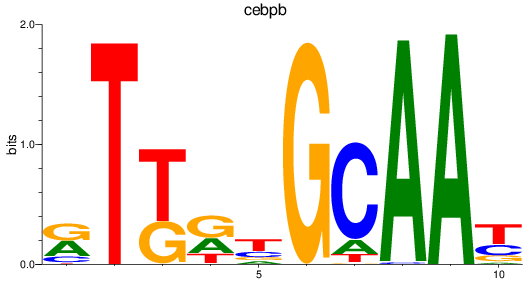
Results for cebpb
Z-value: 2.05
Transcription factors associated with cebpb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
cebpb
|
ENSDARG00000042725 | CCAAT enhancer binding protein beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| cebpb | dr11_v1_chr8_-_28449782_28449782 | 0.47 | 1.6e-06 | Click! |
Activity profile of cebpb motif
Sorted Z-values of cebpb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_23440632 | 59.29 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr5_-_63509581 | 52.99 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr14_+_21106444 | 51.48 |
ENSDART00000075744
ENSDART00000132363 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr14_+_21107032 | 48.36 |
ENSDART00000138319
ENSDART00000139103 ENSDART00000184735 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr9_-_32847642 | 46.68 |
ENSDART00000121506
|
hpx
|
hemopexin |
| chr8_-_50147948 | 42.04 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr13_-_20540790 | 41.58 |
ENSDART00000131467
|
si:ch1073-126c3.2
|
si:ch1073-126c3.2 |
| chr14_+_16345003 | 40.38 |
ENSDART00000003040
ENSDART00000165193 |
itln3
|
intelectin 3 |
| chr1_-_10071422 | 32.84 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr16_-_16701718 | 31.18 |
ENSDART00000143550
|
si:dkey-8k3.2
|
si:dkey-8k3.2 |
| chr20_-_40755614 | 30.97 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr20_+_10538025 | 29.85 |
ENSDART00000129762
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr6_+_22068589 | 28.93 |
ENSDART00000151205
|
aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr22_-_26175237 | 28.08 |
ENSDART00000108737
|
c3b.2
|
complement component c3b, tandem duplicate 2 |
| chr6_+_21202639 | 27.81 |
ENSDART00000083126
|
cidec
|
cell death-inducing DFFA-like effector c |
| chr8_-_39739627 | 27.02 |
ENSDART00000135422
ENSDART00000067844 |
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr3_+_19299309 | 26.32 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr20_+_10544100 | 25.45 |
ENSDART00000113927
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr3_-_40275096 | 25.25 |
ENSDART00000141578
|
shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr19_-_5699703 | 25.06 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr5_+_28830388 | 23.20 |
ENSDART00000149150
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr24_-_33284945 | 23.11 |
ENSDART00000155429
ENSDART00000112845 |
zgc:195173
|
zgc:195173 |
| chr16_-_27749172 | 22.48 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr9_-_9989660 | 22.45 |
ENSDART00000081463
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr22_-_26236188 | 22.01 |
ENSDART00000162640
ENSDART00000167169 ENSDART00000138595 |
c3b.1
|
complement component c3b, tandem duplicate 1 |
| chr7_-_60831082 | 21.95 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr9_-_53062083 | 21.36 |
ENSDART00000122155
|
zmp:0000000936
|
zmp:0000000936 |
| chr5_-_69948099 | 21.03 |
ENSDART00000034639
ENSDART00000191111 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr2_-_32643738 | 20.60 |
ENSDART00000112452
|
si:dkeyp-73d8.9
|
si:dkeyp-73d8.9 |
| chr16_+_23984179 | 19.22 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr9_-_48736388 | 18.97 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr8_+_30699429 | 18.68 |
ENSDART00000005345
|
upb1
|
ureidopropionase, beta |
| chr5_+_26795773 | 18.28 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr7_-_4296771 | 17.01 |
ENSDART00000128855
|
cbln11
|
cerebellin 11 |
| chr9_+_38292947 | 16.80 |
ENSDART00000146663
|
tfcp2l1
|
transcription factor CP2-like 1 |
| chr4_-_77432218 | 16.71 |
ENSDART00000158683
|
slco1d1
|
solute carrier organic anion transporter family, member 1D1 |
| chr16_+_50289916 | 16.27 |
ENSDART00000168861
ENSDART00000167332 |
hamp
|
hepcidin antimicrobial peptide |
| chr14_+_36885524 | 16.05 |
ENSDART00000032547
|
lect2l
|
leukocyte cell-derived chemotaxin 2 like |
| chr25_-_30429607 | 15.81 |
ENSDART00000162429
ENSDART00000176535 |
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr5_-_56412262 | 14.99 |
ENSDART00000083079
|
acaca
|
acetyl-CoA carboxylase alpha |
| chr20_+_37794633 | 14.70 |
ENSDART00000022060
|
atf3
|
activating transcription factor 3 |
| chr16_-_21785261 | 14.63 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr25_+_13191391 | 14.54 |
ENSDART00000109937
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr22_-_17595310 | 14.21 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr18_+_15644559 | 14.11 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr7_+_65240227 | 14.00 |
ENSDART00000168287
|
bco1l
|
beta-carotene oxygenase 1, like |
| chr25_-_32311048 | 13.91 |
ENSDART00000181806
ENSDART00000086334 |
CU372926.1
|
|
| chr15_-_1885247 | 13.76 |
ENSDART00000149703
|
porb
|
P450 (cytochrome) oxidoreductase b |
| chr21_-_39058490 | 13.59 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr2_-_127945 | 13.50 |
ENSDART00000056453
|
igfbp1b
|
insulin-like growth factor binding protein 1b |
| chr15_-_29348212 | 13.46 |
ENSDART00000133117
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr19_+_8606883 | 13.40 |
ENSDART00000054469
ENSDART00000185264 |
s100a10a
|
S100 calcium binding protein A10a |
| chr12_+_48390715 | 13.39 |
ENSDART00000149351
|
scd
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr7_-_7845540 | 13.26 |
ENSDART00000166280
|
cxcl8b.1
|
chemokine (C-X-C motif) ligand 8b, duplicate 1 |
| chr24_-_36727922 | 12.91 |
ENSDART00000135142
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr25_+_13191615 | 12.89 |
ENSDART00000168849
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr16_+_44298902 | 12.81 |
ENSDART00000114795
|
dpys
|
dihydropyrimidinase |
| chr6_+_42475730 | 12.74 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr4_-_9549693 | 12.56 |
ENSDART00000160242
|
FQ377934.1
|
|
| chr4_-_12725513 | 11.66 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr25_+_31323978 | 11.57 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr17_-_15149192 | 11.46 |
ENSDART00000180511
ENSDART00000103405 |
gch1
|
GTP cyclohydrolase 1 |
| chr13_+_33688474 | 11.02 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr12_+_25775734 | 10.82 |
ENSDART00000024415
ENSDART00000149198 |
epas1a
|
endothelial PAS domain protein 1a |
| chr24_-_32582378 | 10.58 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr17_-_6076266 | 10.55 |
ENSDART00000171084
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr5_-_33259079 | 10.36 |
ENSDART00000132223
|
ifitm1
|
interferon induced transmembrane protein 1 |
| chr21_+_4509483 | 10.31 |
ENSDART00000025612
|
phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr1_+_14253118 | 10.31 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr10_+_4924388 | 10.05 |
ENSDART00000108595
|
slc46a2
|
solute carrier family 46 member 2 |
| chr7_-_46019756 | 10.00 |
ENSDART00000162583
|
zgc:162297
|
zgc:162297 |
| chr24_-_32582880 | 9.92 |
ENSDART00000186307
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr12_-_5120175 | 9.83 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr6_-_13498745 | 9.41 |
ENSDART00000027684
ENSDART00000189438 |
mylkb
|
myosin light chain kinase b |
| chr12_-_17199381 | 9.38 |
ENSDART00000193292
|
lipf
|
lipase, gastric |
| chr6_-_37422841 | 9.30 |
ENSDART00000138351
|
cth
|
cystathionase (cystathionine gamma-lyase) |
| chr11_+_43419809 | 9.30 |
ENSDART00000172982
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr8_-_26961779 | 9.16 |
ENSDART00000099214
|
slc16a1b
|
solute carrier family 16 (monocarboxylate transporter), member 1b |
| chr6_-_43449013 | 9.08 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr3_+_3641429 | 9.00 |
ENSDART00000092393
|
plbd1
|
phospholipase B domain containing 1 |
| chr22_-_17677947 | 8.98 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr1_+_17892944 | 8.96 |
ENSDART00000013021
|
tlr3
|
toll-like receptor 3 |
| chr8_+_20438884 | 8.68 |
ENSDART00000016422
ENSDART00000133794 |
mknk2b
|
MAP kinase interacting serine/threonine kinase 2b |
| chr7_-_22790630 | 8.65 |
ENSDART00000173496
|
si:ch211-15b10.6
|
si:ch211-15b10.6 |
| chr7_+_21787507 | 8.58 |
ENSDART00000100936
|
tmem88b
|
transmembrane protein 88 b |
| chr1_-_25177086 | 8.49 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr17_+_10318071 | 8.48 |
ENSDART00000161844
|
foxa1
|
forkhead box A1 |
| chr10_-_36808348 | 8.46 |
ENSDART00000099320
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr17_+_3124129 | 8.44 |
ENSDART00000155323
|
zgc:136872
|
zgc:136872 |
| chr5_+_8196264 | 8.29 |
ENSDART00000174564
ENSDART00000161261 |
lmbrd2a
|
LMBR1 domain containing 2a |
| chr7_+_27691647 | 8.19 |
ENSDART00000079091
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr21_+_20901505 | 8.19 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr17_-_6076084 | 8.16 |
ENSDART00000058890
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr8_-_28349859 | 8.15 |
ENSDART00000062671
|
tuba8l
|
tubulin, alpha 8 like |
| chr20_+_36629173 | 7.91 |
ENSDART00000161241
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr4_-_20181964 | 7.86 |
ENSDART00000022539
|
fgl2a
|
fibrinogen-like 2a |
| chr24_-_32665283 | 7.74 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr19_-_15192840 | 7.48 |
ENSDART00000151337
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr3_-_26204867 | 7.44 |
ENSDART00000103748
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr4_-_19693978 | 7.43 |
ENSDART00000100974
ENSDART00000040405 |
snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr4_+_21866851 | 7.39 |
ENSDART00000177640
|
acss3
|
acyl-CoA synthetase short chain family member 3 |
| chr12_+_46960579 | 7.38 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr7_+_12950507 | 7.31 |
ENSDART00000067629
ENSDART00000158004 |
saa
|
serum amyloid A |
| chr11_-_6265574 | 7.23 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr8_-_19280856 | 7.20 |
ENSDART00000100473
|
zgc:77486
|
zgc:77486 |
| chr19_+_5674907 | 7.12 |
ENSDART00000042189
|
pdk2b
|
pyruvate dehydrogenase kinase, isozyme 2b |
| chr24_-_17049270 | 7.00 |
ENSDART00000175508
|
msrb2
|
methionine sulfoxide reductase B2 |
| chr7_-_7823662 | 6.93 |
ENSDART00000167652
|
cxcl8b.3
|
chemokine (C-X-C motif) ligand 8b, duplicate 3 |
| chr21_+_17051478 | 6.90 |
ENSDART00000047201
ENSDART00000161650 ENSDART00000167298 |
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr18_+_17493859 | 6.78 |
ENSDART00000090754
|
si:dkey-102f14.5
|
si:dkey-102f14.5 |
| chr15_-_17960228 | 6.54 |
ENSDART00000155898
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr7_+_15736230 | 6.49 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr21_-_22715297 | 6.46 |
ENSDART00000065548
|
c1qb
|
complement component 1, q subcomponent, B chain |
| chr12_-_22238004 | 6.42 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr12_+_31616412 | 6.40 |
ENSDART00000124439
|
cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr14_-_38865800 | 6.22 |
ENSDART00000173047
|
gsr
|
glutathione reductase |
| chr17_+_33158350 | 6.22 |
ENSDART00000104476
|
snx9a
|
sorting nexin 9a |
| chr12_+_4920451 | 6.18 |
ENSDART00000171525
ENSDART00000159986 |
plekhm1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr2_-_40199780 | 6.15 |
ENSDART00000113901
|
ccl34a.4
|
chemokine (C-C motif) ligand 34a, duplicate 4 |
| chr10_-_29733194 | 5.98 |
ENSDART00000149252
|
si:ch73-261i21.2
|
si:ch73-261i21.2 |
| chr23_-_29394505 | 5.63 |
ENSDART00000017728
|
pgd
|
phosphogluconate dehydrogenase |
| chr24_-_38644937 | 5.60 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr20_-_33675676 | 5.52 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr9_+_9502610 | 5.44 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr5_-_30615901 | 5.38 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr3_+_3573696 | 5.35 |
ENSDART00000169680
|
CR589947.2
|
|
| chr14_+_48862987 | 5.28 |
ENSDART00000167810
|
zgc:154054
|
zgc:154054 |
| chr21_-_22547496 | 5.19 |
ENSDART00000166835
ENSDART00000089030 |
myo5b
|
myosin VB |
| chr5_-_43935119 | 5.10 |
ENSDART00000142271
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr3_+_19216567 | 4.96 |
ENSDART00000134433
|
il12rb2l
|
interleukin 12 receptor, beta 2a, like |
| chr9_-_9225980 | 4.96 |
ENSDART00000180301
|
cbsb
|
cystathionine-beta-synthase b |
| chr4_-_22335247 | 4.95 |
ENSDART00000192781
|
golgb1
|
golgin B1 |
| chr5_-_6745442 | 4.80 |
ENSDART00000157402
ENSDART00000128684 ENSDART00000168698 |
ostf1
|
osteoclast stimulating factor 1 |
| chr3_-_54524194 | 4.77 |
ENSDART00000155406
ENSDART00000111791 |
si:ch73-208g10.1
|
si:ch73-208g10.1 |
| chr5_-_39474235 | 4.72 |
ENSDART00000171557
|
antxr2a
|
anthrax toxin receptor 2a |
| chr17_+_30450163 | 4.67 |
ENSDART00000104257
|
lpin1
|
lipin 1 |
| chr5_-_32336613 | 4.65 |
ENSDART00000139732
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr2_-_37277626 | 4.64 |
ENSDART00000135340
|
nadkb
|
NAD kinase b |
| chr12_-_45304971 | 4.58 |
ENSDART00000186537
ENSDART00000126405 |
fdxr
|
ferredoxin reductase |
| chr23_+_25305431 | 4.53 |
ENSDART00000143291
|
RPL41
|
si:dkey-151g10.6 |
| chr21_-_16219400 | 4.51 |
ENSDART00000124871
|
tnfrsfa
|
tumor necrosis factor receptor superfamily, member a |
| chr12_-_8070969 | 4.46 |
ENSDART00000020995
|
tmem26b
|
transmembrane protein 26b |
| chr11_-_34577034 | 4.44 |
ENSDART00000133302
ENSDART00000184367 |
pfkfb4a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4a |
| chr18_+_2228737 | 4.44 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr14_-_38929885 | 4.40 |
ENSDART00000148737
|
btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chr14_+_3507326 | 4.36 |
ENSDART00000159326
|
gstp1
|
glutathione S-transferase pi 1 |
| chr20_-_32045057 | 4.26 |
ENSDART00000152970
ENSDART00000034248 |
rab32a
|
RAB32a, member RAS oncogene family |
| chr23_+_35650771 | 4.25 |
ENSDART00000005158
|
ccnt1
|
cyclin T1 |
| chr19_-_33212023 | 4.18 |
ENSDART00000189209
|
trib1
|
tribbles pseudokinase 1 |
| chr3_+_14641962 | 4.17 |
ENSDART00000091070
|
zgc:158403
|
zgc:158403 |
| chr5_+_44346691 | 4.17 |
ENSDART00000034523
|
tars
|
threonyl-tRNA synthetase |
| chr4_-_5595237 | 4.14 |
ENSDART00000109854
|
vegfab
|
vascular endothelial growth factor Ab |
| chr21_-_14762944 | 4.13 |
ENSDART00000114096
|
arrdc1b
|
arrestin domain containing 1b |
| chr8_+_42917515 | 4.07 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr1_+_55752593 | 4.05 |
ENSDART00000108838
ENSDART00000134770 |
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr2_+_17055069 | 4.02 |
ENSDART00000115078
|
thpo
|
thrombopoietin |
| chr11_+_37251825 | 3.97 |
ENSDART00000169804
|
il17rc
|
interleukin 17 receptor C |
| chr23_+_30967686 | 3.94 |
ENSDART00000144485
|
si:ch211-197l9.2
|
si:ch211-197l9.2 |
| chr13_-_31544365 | 3.94 |
ENSDART00000005670
|
dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr1_-_52222989 | 3.84 |
ENSDART00000010236
|
cnn1a
|
calponin 1, basic, smooth muscle, a |
| chr2_+_11029138 | 3.79 |
ENSDART00000138737
ENSDART00000081058 ENSDART00000153662 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr20_+_25626479 | 3.66 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr17_+_25519089 | 3.66 |
ENSDART00000041721
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr24_-_27436319 | 3.65 |
ENSDART00000171489
|
si:dkey-25o1.7
|
si:dkey-25o1.7 |
| chr1_-_9486214 | 3.61 |
ENSDART00000137821
|
micall2b
|
mical-like 2b |
| chr8_+_21229718 | 3.60 |
ENSDART00000100222
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr12_-_33706726 | 3.46 |
ENSDART00000153135
|
myo15b
|
myosin XVB |
| chr25_+_35553542 | 3.44 |
ENSDART00000113723
|
spi1a
|
Spi-1 proto-oncogene a |
| chr24_+_16983269 | 3.43 |
ENSDART00000139176
ENSDART00000023833 |
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr10_+_23099890 | 3.43 |
ENSDART00000135890
|
LTN1
|
si:dkey-175g6.5 |
| chr5_-_43935460 | 3.42 |
ENSDART00000166152
ENSDART00000188969 |
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr21_-_30111134 | 3.41 |
ENSDART00000014223
|
slc23a1
|
solute carrier family 23 (ascorbic acid transporter), member 1 |
| chr17_-_42799104 | 3.37 |
ENSDART00000154755
|
prkd3
|
protein kinase D3 |
| chr19_-_34927201 | 3.37 |
ENSDART00000076518
|
sla1
|
Src-like-adaptor 1 |
| chr22_+_15973122 | 3.34 |
ENSDART00000144545
|
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr17_+_10501647 | 3.32 |
ENSDART00000140391
|
tyro3
|
TYRO3 protein tyrosine kinase |
| chr6_+_8314451 | 3.31 |
ENSDART00000147793
ENSDART00000183688 |
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr13_+_41917606 | 3.31 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr16_-_51253925 | 3.30 |
ENSDART00000050644
|
serpinb14
|
serpin peptidase inhibitor, clade B (ovalbumin), member 14 |
| chr9_+_907459 | 3.29 |
ENSDART00000034850
ENSDART00000144114 |
dbi
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr7_+_38090515 | 3.16 |
ENSDART00000131387
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr14_-_1098607 | 3.16 |
ENSDART00000169090
ENSDART00000158905 |
FQ311908.1
|
|
| chr12_+_46462090 | 3.13 |
ENSDART00000130748
|
BX005305.2
|
|
| chr1_-_59126139 | 3.09 |
ENSDART00000156105
|
si:ch1073-110a20.7
|
si:ch1073-110a20.7 |
| chr15_-_17010358 | 3.09 |
ENSDART00000156768
|
hip1
|
huntingtin interacting protein 1 |
| chr3_+_37649436 | 3.08 |
ENSDART00000075039
|
gosr2
|
golgi SNAP receptor complex member 2 |
| chr5_-_26181863 | 3.05 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr23_-_25891426 | 3.02 |
ENSDART00000189907
|
CR318614.1
|
|
| chr16_-_32233463 | 2.99 |
ENSDART00000102016
|
calhm6
|
calcium homeostasis modulator family member 6 |
| chr1_-_18585046 | 2.99 |
ENSDART00000147228
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr20_-_25533739 | 2.98 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr6_+_39114345 | 2.98 |
ENSDART00000136835
|
gpr84
|
G protein-coupled receptor 84 |
| chr8_-_23599096 | 2.96 |
ENSDART00000183096
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr22_-_13165186 | 2.90 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr16_+_27442549 | 2.90 |
ENSDART00000015688
|
invs
|
inversin |
| chr3_-_57744323 | 2.90 |
ENSDART00000101829
|
lgals3bpb
|
lectin, galactoside-binding, soluble, 3 binding protein b |
| chr22_-_10397600 | 2.83 |
ENSDART00000181964
ENSDART00000142886 |
nisch
|
nischarin |
| chr20_+_32118559 | 2.82 |
ENSDART00000026273
|
cd164
|
CD164 molecule, sialomucin |
| chr18_-_26797723 | 2.78 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr10_-_23099809 | 2.77 |
ENSDART00000148333
ENSDART00000079703 ENSDART00000162444 |
nle1
|
notchless homolog 1 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of cebpb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.6 | 28.9 | GO:0009397 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 8.4 | 25.3 | GO:0006565 | L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 6.1 | 18.3 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 5.6 | 22.5 | GO:0015677 | copper ion import(GO:0015677) |
| 4.4 | 26.3 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 4.3 | 102.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 3.9 | 23.6 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 3.7 | 15.0 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 3.7 | 18.7 | GO:0019482 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 3.4 | 13.5 | GO:0003418 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 3.3 | 13.4 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 3.0 | 9.0 | GO:0034138 | chemokine production(GO:0032602) toll-like receptor 3 signaling pathway(GO:0034138) |
| 2.9 | 11.5 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 2.5 | 7.5 | GO:0015882 | L-ascorbic acid transport(GO:0015882) |
| 2.4 | 7.3 | GO:0006953 | acute-phase response(GO:0006953) |
| 2.4 | 7.2 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 2.3 | 16.3 | GO:0032570 | response to progesterone(GO:0032570) |
| 2.2 | 32.8 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 2.2 | 41.4 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 2.2 | 6.5 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 2.0 | 14.3 | GO:0019344 | homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 1.9 | 7.7 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 1.6 | 8.2 | GO:0071295 | cellular response to nutrient(GO:0031670) cellular response to vitamin(GO:0071295) cellular response to vitamin D(GO:0071305) |
| 1.6 | 6.4 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 1.5 | 9.3 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 1.4 | 9.8 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 1.3 | 5.2 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 1.3 | 23.1 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 1.2 | 13.5 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 1.2 | 22.0 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 1.1 | 4.5 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 1.1 | 9.0 | GO:0030104 | water homeostasis(GO:0030104) |
| 1.1 | 7.5 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 1.0 | 5.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 1.0 | 19.2 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 1.0 | 86.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 1.0 | 4.0 | GO:0030091 | protein repair(GO:0030091) |
| 1.0 | 9.9 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.9 | 8.1 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.9 | 18.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.9 | 14.1 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.9 | 3.4 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.9 | 12.8 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.8 | 8.5 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.7 | 3.0 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.7 | 13.2 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.7 | 2.9 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.7 | 7.2 | GO:0010890 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.7 | 18.7 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.7 | 12.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.7 | 4.6 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.7 | 5.9 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.6 | 5.6 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.6 | 3.4 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.6 | 1.7 | GO:1902001 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) positive regulation of lipid transport(GO:0032370) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.5 | 6.9 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.5 | 4.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.4 | 6.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.4 | 2.0 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.4 | 1.6 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.4 | 1.6 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.4 | 6.8 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.4 | 9.8 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.4 | 4.1 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.4 | 13.6 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.4 | 2.6 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.4 | 11.7 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.3 | 1.0 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.3 | 3.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.3 | 4.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 7.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.3 | 2.8 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.3 | 0.9 | GO:2000726 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.3 | 5.5 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.3 | 8.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.3 | 3.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.3 | 2.7 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.3 | 6.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.3 | 1.8 | GO:0060143 | positive regulation of syncytium formation by plasma membrane fusion(GO:0060143) positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 4.6 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 2.0 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.2 | 0.4 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.2 | 9.1 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.2 | 0.9 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.2 | 4.5 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.2 | 1.0 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.2 | 4.2 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.2 | 1.0 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.2 | 14.7 | GO:0009615 | response to virus(GO:0009615) |
| 0.2 | 4.2 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.2 | 7.1 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.2 | 1.3 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.2 | 8.4 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.2 | 3.4 | GO:0002761 | regulation of myeloid leukocyte differentiation(GO:0002761) |
| 0.1 | 1.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 2.7 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 6.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 6.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 9.4 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.1 | 8.6 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 5.3 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 6.2 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.1 | 4.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.4 | GO:0050847 | osteoclast development(GO:0036035) progesterone receptor signaling pathway(GO:0050847) bone cell development(GO:0098751) |
| 0.1 | 7.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 4.0 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 9.2 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 4.4 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.1 | 2.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 3.3 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 4.7 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.1 | 3.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 1.1 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 4.4 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.1 | 8.6 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.1 | 3.0 | GO:0032496 | response to lipopolysaccharide(GO:0032496) |
| 0.1 | 3.3 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.1 | 8.1 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.1 | 2.4 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.1 | 4.2 | GO:0042493 | response to drug(GO:0042493) |
| 0.1 | 0.3 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 0.5 | GO:0050927 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.1 | 2.5 | GO:0030149 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.1 | 5.0 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.1 | 5.3 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 1.8 | GO:0003014 | renal system process(GO:0003014) |
| 0.1 | 0.6 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.1 | 0.7 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 11.3 | GO:0009725 | response to hormone(GO:0009725) |
| 0.1 | 0.7 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 4.3 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 2.1 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 1.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 4.5 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 2.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 7.5 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.6 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 6.2 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 1.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.4 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 4.2 | GO:0001889 | liver development(GO:0001889) |
| 0.0 | 0.3 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 1.0 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 1.4 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 1.4 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 9.6 | GO:0042981 | regulation of apoptotic process(GO:0042981) |
| 0.0 | 0.4 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 1.7 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 2.7 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 12.7 | GO:0006508 | proteolysis(GO:0006508) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.9 | 32.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.9 | 19.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 1.6 | 6.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.9 | 7.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.9 | 2.7 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.9 | 2.6 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.8 | 3.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.7 | 7.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.7 | 2.8 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.7 | 3.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.6 | 12.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.5 | 4.7 | GO:0070938 | contractile ring(GO:0070938) |
| 0.5 | 8.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.5 | 2.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.5 | 4.2 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.3 | 1.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.3 | 3.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.3 | 4.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.3 | 6.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.3 | 344.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.2 | 6.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 103.0 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.2 | 2.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 2.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 19.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 6.6 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 2.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 7.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.6 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 5.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 24.2 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 1.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 3.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 13.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 9.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 1.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 7.2 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 2.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 27.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 6.2 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 5.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 4.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.3 | GO:0030669 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 11.9 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 21.0 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 2.2 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 4.5 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.5 | 99.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 9.6 | 28.9 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 8.4 | 25.3 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 7.6 | 30.5 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 7.3 | 22.0 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 6.2 | 18.7 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 5.6 | 22.5 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 4.4 | 17.5 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 4.3 | 21.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 4.2 | 16.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 4.1 | 16.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 3.7 | 15.0 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 3.4 | 13.8 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 3.3 | 13.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 2.8 | 14.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 2.6 | 7.9 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 2.4 | 19.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 2.4 | 14.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 2.3 | 18.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 1.8 | 40.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 1.7 | 7.0 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 1.6 | 6.3 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 1.6 | 14.0 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 1.4 | 9.8 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 1.4 | 4.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 1.4 | 13.6 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 1.3 | 5.1 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 1.2 | 6.2 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 1.2 | 6.2 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 1.2 | 13.5 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 1.1 | 189.5 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 1.1 | 12.8 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 1.0 | 4.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 1.0 | 9.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 1.0 | 12.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 1.0 | 5.0 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 1.0 | 56.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.9 | 7.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.8 | 8.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.8 | 4.1 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.8 | 9.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.7 | 5.5 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.6 | 2.5 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.5 | 3.8 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.5 | 13.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.5 | 3.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.4 | 1.6 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.4 | 6.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.4 | 7.5 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.4 | 9.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.4 | 7.7 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.4 | 13.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.4 | 2.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.4 | 16.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 1.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.3 | 2.7 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 4.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 4.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 9.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.3 | 1.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 4.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.3 | 12.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.3 | 3.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.3 | 7.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 8.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 3.0 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.2 | 6.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 1.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 2.3 | GO:0008443 | 6-phosphofructokinase activity(GO:0003872) phosphofructokinase activity(GO:0008443) |
| 0.2 | 3.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 1.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 3.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 11.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.2 | 39.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 9.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.2 | 1.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 1.7 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.2 | 0.8 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 4.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 9.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 3.1 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 8.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 13.9 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 3.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 7.4 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 5.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 1.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 1.4 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 1.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.7 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 5.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 3.4 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 2.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.1 | 0.6 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 13.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 5.3 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 0.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.6 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) |
| 0.1 | 0.4 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 4.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 2.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 5.0 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 5.8 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 3.6 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 2.1 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 7.6 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 3.3 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 2.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 2.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 4.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 1.0 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 5.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.9 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 1.7 | GO:0042626 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 1.4 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.7 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 108.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 1.9 | 32.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.9 | 25.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.8 | 11.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.7 | 53.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.7 | 14.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.5 | 9.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.4 | 16.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.4 | 21.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.3 | 4.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 3.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 23.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 2.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 7.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 3.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 7.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.3 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 3.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 99.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 3.9 | 31.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 2.7 | 32.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 2.3 | 59.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 2.3 | 9.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 1.9 | 19.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 1.6 | 11.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 1.1 | 16.7 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 1.0 | 9.0 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.8 | 4.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.7 | 16.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.7 | 9.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.6 | 7.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.6 | 32.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.5 | 9.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.4 | 25.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.4 | 4.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.4 | 4.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.4 | 10.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.3 | 7.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.3 | 6.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 8.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.3 | 2.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.2 | 4.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 1.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 1.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 1.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 3.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.2 | 4.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 4.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 5.2 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 4.0 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.1 | 1.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 5.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 3.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 3.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 4.2 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |