Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
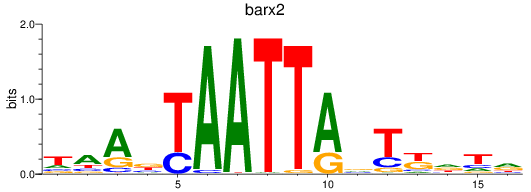
Results for barx2
Z-value: 1.24
Transcription factors associated with barx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
barx2
|
ENSDARG00000041098 | BARX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| barx2 | dr11_v1_chr18_+_47313899_47313899 | 0.04 | 7.2e-01 | Click! |
Activity profile of barx2 motif
Sorted Z-values of barx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_9669717 | 27.86 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr11_+_30057762 | 21.46 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr22_-_13851297 | 19.11 |
ENSDART00000080306
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr16_+_5774977 | 16.18 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr2_-_44280061 | 15.65 |
ENSDART00000136818
|
mpz
|
myelin protein zero |
| chr23_+_40460333 | 14.42 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr8_+_31717175 | 14.31 |
ENSDART00000013434
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr9_-_9348058 | 13.93 |
ENSDART00000132257
|
zgc:113337
|
zgc:113337 |
| chr15_-_44512461 | 12.45 |
ENSDART00000155456
|
gria4a
|
glutamate receptor, ionotropic, AMPA 4a |
| chr25_+_37126921 | 11.90 |
ENSDART00000124331
|
si:ch1073-174d20.1
|
si:ch1073-174d20.1 |
| chr25_-_19420949 | 11.70 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr12_-_10300101 | 11.65 |
ENSDART00000126428
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| chr2_+_20430366 | 11.60 |
ENSDART00000155108
|
si:ch211-153l6.6
|
si:ch211-153l6.6 |
| chr6_-_11768198 | 11.47 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr16_+_46111849 | 11.46 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr5_+_63668735 | 11.06 |
ENSDART00000134261
ENSDART00000097330 |
dnm1b
|
dynamin 1b |
| chr8_+_31716872 | 10.79 |
ENSDART00000161121
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr13_-_29420885 | 10.17 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr18_+_24919614 | 9.92 |
ENSDART00000008638
|
rgma
|
repulsive guidance molecule family member a |
| chr17_-_20717845 | 9.66 |
ENSDART00000150037
|
ank3b
|
ankyrin 3b |
| chr16_-_28856112 | 9.58 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr20_+_41756996 | 9.55 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr23_-_26521970 | 9.34 |
ENSDART00000143712
|
si:dkey-205h13.1
|
si:dkey-205h13.1 |
| chr21_-_20939488 | 9.34 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr19_-_26869103 | 9.13 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr2_-_30784198 | 8.90 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr12_+_18578597 | 8.83 |
ENSDART00000134944
|
grid2ipb
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, b |
| chr16_-_25568512 | 8.59 |
ENSDART00000149411
|
atxn1b
|
ataxin 1b |
| chr9_-_42418470 | 8.46 |
ENSDART00000144353
|
calcrla
|
calcitonin receptor-like a |
| chr11_+_30663300 | 8.38 |
ENSDART00000161662
|
ttbk1a
|
tau tubulin kinase 1a |
| chr16_+_54209504 | 8.32 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr1_-_19502322 | 8.31 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr6_-_40744720 | 8.19 |
ENSDART00000154916
ENSDART00000186922 |
p4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr18_-_48983690 | 8.09 |
ENSDART00000182359
|
FO681288.3
|
|
| chr5_+_4054704 | 7.94 |
ENSDART00000140537
|
dhrs11a
|
dehydrogenase/reductase (SDR family) member 11a |
| chr13_+_7292061 | 7.92 |
ENSDART00000179504
|
CABZ01072077.1
|
Danio rerio neuroblast differentiation-associated protein AHNAK-like (LOC795051), mRNA. |
| chr4_-_2219705 | 7.88 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr1_-_50859053 | 7.71 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr12_+_4971515 | 7.64 |
ENSDART00000161076
|
arhgap27
|
Rho GTPase activating protein 27 |
| chr12_+_1139690 | 7.57 |
ENSDART00000160442
|
CABZ01072885.1
|
|
| chr6_-_6487876 | 7.55 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr7_-_25895189 | 7.37 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr16_-_12173554 | 7.29 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr3_-_20040636 | 7.24 |
ENSDART00000104118
|
atxn7l3
|
ataxin 7-like 3 |
| chr5_-_6377865 | 7.12 |
ENSDART00000031775
|
zgc:73226
|
zgc:73226 |
| chr4_-_5019113 | 6.91 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr10_-_21054059 | 6.75 |
ENSDART00000139733
|
pcdh1a
|
protocadherin 1a |
| chr6_-_30210378 | 6.72 |
ENSDART00000157359
ENSDART00000113924 |
lrrc7
|
leucine rich repeat containing 7 |
| chr21_-_39177564 | 6.69 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr7_-_49594995 | 6.68 |
ENSDART00000174161
ENSDART00000109147 |
brsk2b
|
BR serine/threonine kinase 2b |
| chr3_+_46459540 | 6.54 |
ENSDART00000188150
|
si:ch211-66e2.5
|
si:ch211-66e2.5 |
| chr24_-_28437833 | 6.47 |
ENSDART00000125412
|
fbxo45
|
F-box protein 45 |
| chr15_-_755023 | 6.38 |
ENSDART00000155594
|
znf1011
|
zinc finger protein 1011 |
| chr6_-_29305132 | 6.18 |
ENSDART00000132456
|
bivm
|
basic, immunoglobulin-like variable motif containing |
| chr24_+_36204028 | 6.04 |
ENSDART00000063832
ENSDART00000155260 |
rbbp8
|
retinoblastoma binding protein 8 |
| chr15_+_15856178 | 6.03 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr9_+_7998794 | 6.02 |
ENSDART00000138167
|
myo16
|
myosin XVI |
| chr24_-_15131831 | 5.92 |
ENSDART00000028410
|
cd226
|
CD226 molecule |
| chr25_-_27722614 | 5.84 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr21_+_13387965 | 5.82 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr22_+_18816662 | 5.67 |
ENSDART00000132476
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr17_-_2386569 | 5.64 |
ENSDART00000121614
|
PLCB2
|
phospholipase C beta 2 |
| chr22_-_26865361 | 5.63 |
ENSDART00000182504
|
hmox2a
|
heme oxygenase 2a |
| chr8_-_14375890 | 5.55 |
ENSDART00000090306
|
xpr1a
|
xenotropic and polytropic retrovirus receptor 1a |
| chr15_+_15516612 | 5.51 |
ENSDART00000016024
|
traf4a
|
tnf receptor-associated factor 4a |
| chr25_+_16116740 | 5.48 |
ENSDART00000139778
|
far1
|
fatty acyl CoA reductase 1 |
| chr4_+_4849789 | 5.47 |
ENSDART00000130818
ENSDART00000127751 |
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr23_+_4253957 | 5.44 |
ENSDART00000008761
|
arl6ip5b
|
ADP-ribosylation factor-like 6 interacting protein 5b |
| chr9_+_54039006 | 5.44 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr23_+_17899226 | 5.36 |
ENSDART00000079487
|
CU234171.1
|
|
| chr7_+_20629411 | 5.35 |
ENSDART00000173710
|
si:dkey-19b23.15
|
si:dkey-19b23.15 |
| chr5_-_14373662 | 5.28 |
ENSDART00000183694
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr2_+_11923615 | 5.20 |
ENSDART00000126118
|
trove2
|
TROVE domain family, member 2 |
| chr4_+_17279966 | 5.19 |
ENSDART00000067005
ENSDART00000137487 |
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr24_+_28953089 | 5.18 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr6_+_13933464 | 5.08 |
ENSDART00000109144
|
ptprnb
|
protein tyrosine phosphatase, receptor type, Nb |
| chr20_+_19512727 | 5.07 |
ENSDART00000063696
|
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr21_+_13383413 | 4.99 |
ENSDART00000151345
|
zgc:113162
|
zgc:113162 |
| chr5_+_65536095 | 4.98 |
ENSDART00000189898
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr7_+_19552381 | 4.94 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr2_-_23004286 | 4.94 |
ENSDART00000134664
ENSDART00000110373 ENSDART00000185833 ENSDART00000187235 |
znf414
mllt1b
|
zinc finger protein 414 MLLT1, super elongation complex subunit b |
| chr5_-_66702479 | 4.92 |
ENSDART00000129197
|
mn1b
|
meningioma 1b |
| chr5_+_63302660 | 4.90 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr16_+_40024883 | 4.83 |
ENSDART00000110100
|
hint3
|
histidine triad nucleotide binding protein 3 |
| chr7_-_48805181 | 4.83 |
ENSDART00000015884
|
mfge8a
|
milk fat globule-EGF factor 8 protein a |
| chr2_-_3678029 | 4.82 |
ENSDART00000146861
|
mmp16b
|
matrix metallopeptidase 16b (membrane-inserted) |
| chr17_-_11329959 | 4.75 |
ENSDART00000015418
|
irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr21_-_28340977 | 4.73 |
ENSDART00000141629
|
nrxn2a
|
neurexin 2a |
| chr18_-_30020879 | 4.68 |
ENSDART00000162086
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr14_-_470505 | 4.63 |
ENSDART00000067147
|
ANKRD50
|
ankyrin repeat domain 50 |
| chr20_+_34717403 | 4.57 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr20_+_40457599 | 4.57 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr25_-_29988352 | 4.56 |
ENSDART00000067059
|
fam19a5b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5b |
| chr8_-_50888806 | 4.53 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr16_-_35329803 | 4.52 |
ENSDART00000161729
ENSDART00000157700 ENSDART00000184584 ENSDART00000174713 ENSDART00000162518 |
ptprub
|
protein tyrosine phosphatase, receptor type, U, b |
| chr1_-_22512063 | 4.44 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr3_-_48716422 | 4.43 |
ENSDART00000164979
|
si:ch211-114m9.1
|
si:ch211-114m9.1 |
| chr16_+_22761846 | 4.40 |
ENSDART00000193028
|
CR854942.1
|
|
| chr1_+_40107811 | 4.39 |
ENSDART00000113954
|
si:ch211-113e8.11
|
si:ch211-113e8.11 |
| chr11_+_29770966 | 4.39 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr3_+_62339264 | 4.38 |
ENSDART00000174569
|
BX470259.3
|
|
| chr16_-_44945224 | 4.37 |
ENSDART00000156921
|
ncam3
|
neural cell adhesion molecule 3 |
| chr5_-_12407194 | 4.37 |
ENSDART00000125291
|
ksr2
|
kinase suppressor of ras 2 |
| chr3_+_60721342 | 4.28 |
ENSDART00000157772
|
foxj1a
|
forkhead box J1a |
| chr23_-_36446307 | 4.27 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr13_-_12494575 | 4.24 |
ENSDART00000137761
|
si:dkey-20i10.7
|
si:dkey-20i10.7 |
| chr11_+_11267493 | 4.23 |
ENSDART00000148425
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr10_-_7386475 | 4.23 |
ENSDART00000167963
|
nrg1
|
neuregulin 1 |
| chr25_-_25058508 | 4.20 |
ENSDART00000087570
ENSDART00000178891 |
FQ311928.1
|
|
| chr3_+_24190207 | 4.19 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr22_+_1700552 | 4.15 |
ENSDART00000166185
|
znf1154
|
zinc finger protein 1154 |
| chr6_-_40466861 | 4.14 |
ENSDART00000062724
|
crbn
|
cereblon |
| chr15_-_9257136 | 4.14 |
ENSDART00000187901
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr10_+_42733210 | 4.10 |
ENSDART00000189832
|
CABZ01063556.1
|
|
| chr12_+_8168272 | 4.08 |
ENSDART00000054092
|
arid5b
|
AT-rich interaction domain 5B |
| chr3_+_24537023 | 4.05 |
ENSDART00000077702
|
sp100.1
|
SP110 nuclear body protein, tandem duplicate 1 |
| chr2_+_57848844 | 4.01 |
ENSDART00000037279
|
plekhj1
|
pleckstrin homology domain containing, family J member 1 |
| chr18_-_215698 | 4.01 |
ENSDART00000147430
|
tarsl2
|
threonyl-tRNA synthetase-like 2 |
| chr1_+_51312752 | 4.00 |
ENSDART00000063938
|
mast1a
|
microtubule associated serine/threonine kinase 1a |
| chr20_-_53078607 | 3.94 |
ENSDART00000163494
ENSDART00000191730 |
CABZ01066813.1
|
|
| chr22_+_2769236 | 3.90 |
ENSDART00000141836
|
si:dkey-20i20.10
|
si:dkey-20i20.10 |
| chr1_+_45663727 | 3.90 |
ENSDART00000038574
ENSDART00000141144 ENSDART00000149565 |
trappc5
|
trafficking protein particle complex 5 |
| chr12_-_35885349 | 3.89 |
ENSDART00000085662
|
cep131
|
centrosomal protein 131 |
| chr14_-_16082806 | 3.88 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr2_-_42065069 | 3.86 |
ENSDART00000140188
|
cspp1b
|
centrosome and spindle pole associated protein 1b |
| chr23_+_7710447 | 3.84 |
ENSDART00000168199
|
kif3b
|
kinesin family member 3B |
| chr6_+_49551614 | 3.83 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr10_-_36927594 | 3.80 |
ENSDART00000162046
|
sez6a
|
seizure related 6 homolog a |
| chr17_-_39772999 | 3.80 |
ENSDART00000155727
|
pimr60
|
Pim proto-oncogene, serine/threonine kinase, related 60 |
| chr9_+_2574122 | 3.77 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr23_+_32021803 | 3.58 |
ENSDART00000012963
|
trappc6b
|
trafficking protein particle complex 6b |
| chr17_-_16422654 | 3.58 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr13_+_15656042 | 3.56 |
ENSDART00000134240
|
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr7_-_40145097 | 3.54 |
ENSDART00000173634
|
wdr60
|
WD repeat domain 60 |
| chr1_-_15797663 | 3.53 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr17_+_41992054 | 3.52 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr7_-_71829649 | 3.51 |
ENSDART00000160449
|
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr1_-_22757145 | 3.44 |
ENSDART00000134719
|
prom1b
|
prominin 1 b |
| chr10_+_45089820 | 3.44 |
ENSDART00000175481
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr1_+_513986 | 3.44 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr16_+_28994709 | 3.43 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr3_+_57268363 | 3.42 |
ENSDART00000180725
|
TMEM235 (1 of many)
|
transmembrane protein 235 |
| chr21_+_38732945 | 3.40 |
ENSDART00000076157
|
rab24
|
RAB24, member RAS oncogene family |
| chr18_-_30021479 | 3.35 |
ENSDART00000144562
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr21_+_27513859 | 3.33 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr10_+_15967643 | 3.32 |
ENSDART00000136709
|
apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr16_-_12173399 | 3.31 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr8_+_7097740 | 3.30 |
ENSDART00000159670
|
abtb1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr11_-_1509773 | 3.30 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr3_+_62205858 | 3.29 |
ENSDART00000126807
|
zgc:173575
|
zgc:173575 |
| chr5_-_37881345 | 3.29 |
ENSDART00000084819
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr22_-_17844117 | 3.27 |
ENSDART00000159363
|
BX908731.1
|
|
| chr8_+_44722140 | 3.24 |
ENSDART00000163381
|
elmod3
|
ELMO/CED-12 domain containing 3 |
| chr11_+_42726712 | 3.23 |
ENSDART00000028955
|
tdrd3
|
tudor domain containing 3 |
| chr3_-_26806032 | 3.22 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr10_-_29236860 | 3.21 |
ENSDART00000111620
|
ccdc83
|
coiled-coil domain containing 83 |
| chr8_+_23521974 | 3.20 |
ENSDART00000188130
ENSDART00000129378 |
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr1_+_55293424 | 3.16 |
ENSDART00000152464
|
zgc:172106
|
zgc:172106 |
| chr12_+_48480632 | 3.14 |
ENSDART00000158157
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr2_+_7295515 | 3.14 |
ENSDART00000152987
|
si:dkeyp-106c3.1
|
si:dkeyp-106c3.1 |
| chr4_+_2482046 | 3.14 |
ENSDART00000103371
|
zdhhc17
|
zinc finger, DHHC-type containing 17 |
| chr1_+_38775041 | 3.12 |
ENSDART00000110824
|
wdr17
|
WD repeat domain 17 |
| chr2_-_33645411 | 3.10 |
ENSDART00000114663
|
b4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr21_+_723998 | 3.10 |
ENSDART00000160956
|
oaz1b
|
ornithine decarboxylase antizyme 1b |
| chr3_+_62271320 | 3.08 |
ENSDART00000132197
|
BX470259.2
|
|
| chr5_-_41494831 | 3.07 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr1_+_6862917 | 3.04 |
ENSDART00000182953
|
erbb4a
|
erb-b2 receptor tyrosine kinase 4a |
| chr10_-_7857494 | 3.03 |
ENSDART00000143215
|
inpp5ja
|
inositol polyphosphate-5-phosphatase Ja |
| chr11_+_31323746 | 3.02 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr22_+_30496360 | 3.01 |
ENSDART00000189145
|
BX649490.4
|
|
| chr16_+_25074029 | 2.98 |
ENSDART00000155465
|
si:dkeyp-84f3.9
|
si:dkeyp-84f3.9 |
| chr16_-_22781446 | 2.92 |
ENSDART00000144107
|
lenep
|
lens epithelial protein |
| chr1_+_11977426 | 2.86 |
ENSDART00000103399
|
tspan5b
|
tetraspanin 5b |
| chr24_+_12983434 | 2.85 |
ENSDART00000145214
ENSDART00000146911 ENSDART00000066700 |
eloca
|
elongin C paralog a |
| chr21_-_2232640 | 2.85 |
ENSDART00000157754
|
zgc:113343
|
zgc:113343 |
| chr18_+_17725410 | 2.84 |
ENSDART00000090608
|
rspry1
|
ring finger and SPRY domain containing 1 |
| chr12_+_14556092 | 2.84 |
ENSDART00000115237
|
becn1
|
beclin 1, autophagy related |
| chr13_-_38730267 | 2.83 |
ENSDART00000157524
|
lmbrd1
|
LMBR1 domain containing 1 |
| chr7_+_19528265 | 2.82 |
ENSDART00000005326
|
ttc5
|
tetratricopeptide repeat domain 5 |
| chr24_-_7995960 | 2.80 |
ENSDART00000186594
|
bloc1s5
|
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr15_-_40267485 | 2.79 |
ENSDART00000152253
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr24_-_29586082 | 2.79 |
ENSDART00000136763
|
vav3a
|
vav 3 guanine nucleotide exchange factor a |
| chr19_-_3931917 | 2.78 |
ENSDART00000162532
|
map7d1b
|
MAP7 domain containing 1b |
| chr23_+_36460239 | 2.77 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr23_-_36303216 | 2.77 |
ENSDART00000188720
|
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr15_-_14552101 | 2.77 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr16_-_36748374 | 2.72 |
ENSDART00000133310
|
pik3r4
|
phosphoinositide-3-kinase, regulatory subunit 4 |
| chr17_+_49281597 | 2.72 |
ENSDART00000155599
|
zgc:113176
|
zgc:113176 |
| chr19_-_42045372 | 2.69 |
ENSDART00000144275
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr21_+_22840246 | 2.67 |
ENSDART00000151621
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr23_+_33991509 | 2.65 |
ENSDART00000145161
|
fam50a
|
family with sequence similarity 50, member A |
| chr10_+_26652859 | 2.64 |
ENSDART00000079174
|
htatsf1
|
HIV-1 Tat specific factor 1 |
| chr8_+_28358161 | 2.63 |
ENSDART00000062682
|
adipor1b
|
adiponectin receptor 1b |
| chr15_-_18162647 | 2.61 |
ENSDART00000012064
|
pih1d2
|
PIH1 domain containing 2 |
| chr5_-_55933420 | 2.61 |
ENSDART00000050966
|
slc25a46
|
solute carrier family 25, member 46 |
| chr3_-_30488063 | 2.59 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr22_-_18179214 | 2.58 |
ENSDART00000129576
|
si:ch211-125m10.6
|
si:ch211-125m10.6 |
| chr15_-_47865063 | 2.58 |
ENSDART00000151600
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr11_+_24800156 | 2.57 |
ENSDART00000131976
|
adipor1a
|
adiponectin receptor 1a |
| chr4_+_77047067 | 2.56 |
ENSDART00000174206
|
znf1009
|
zinc finger protein 1009 |
Network of associatons between targets according to the STRING database.
First level regulatory network of barx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 25.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 3.4 | 10.2 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 2.4 | 11.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 2.1 | 8.3 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 2.0 | 6.0 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 2.0 | 9.9 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 2.0 | 5.9 | GO:0002836 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 1.8 | 5.4 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 1.4 | 4.2 | GO:0060945 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 1.4 | 5.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 1.3 | 5.2 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 1.3 | 5.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 1.2 | 10.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 1.1 | 4.4 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 1.1 | 5.3 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.9 | 2.8 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.9 | 5.6 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.9 | 4.3 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.8 | 6.7 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.8 | 5.7 | GO:1903306 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.8 | 7.1 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.8 | 5.4 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.7 | 6.5 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.7 | 7.9 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.7 | 2.8 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.7 | 5.4 | GO:0043584 | nose development(GO:0043584) |
| 0.7 | 2.7 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.7 | 2.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.6 | 2.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.6 | 7.0 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.6 | 11.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.6 | 3.9 | GO:0032387 | negative regulation of intracellular transport(GO:0032387) intraciliary transport involved in cilium morphogenesis(GO:0035735) negative regulation of intracellular protein transport(GO:0090317) |
| 0.5 | 3.1 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.5 | 2.0 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.5 | 1.5 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.5 | 6.7 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.5 | 6.9 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.5 | 2.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.5 | 11.7 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.4 | 1.3 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.4 | 8.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.4 | 1.3 | GO:1903430 | negative regulation of cell maturation(GO:1903430) |
| 0.4 | 1.6 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.4 | 2.5 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.4 | 4.1 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.4 | 8.5 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.4 | 3.0 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.4 | 4.9 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.4 | 2.6 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.4 | 16.2 | GO:0007586 | digestion(GO:0007586) |
| 0.3 | 2.0 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.3 | 1.0 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.3 | 4.0 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.3 | 1.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.3 | 12.3 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.3 | 1.5 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.3 | 2.6 | GO:0090148 | membrane fission(GO:0090148) |
| 0.3 | 2.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.3 | 1.4 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.3 | 15.7 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.3 | 4.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.3 | 1.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 1.9 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.2 | 2.3 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.2 | 6.1 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.2 | 2.0 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.2 | 2.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.2 | 2.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 1.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.2 | 3.1 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.2 | 1.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 7.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.2 | 2.0 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.2 | 5.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.2 | 5.6 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.2 | 3.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.2 | 2.4 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 1.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 5.8 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.2 | 9.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.2 | 2.5 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.2 | 1.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.2 | 1.1 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.2 | 6.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.2 | 5.8 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 16.9 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 4.8 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 1.4 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 2.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 2.0 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 2.6 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 15.9 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.1 | 2.2 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 2.5 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 3.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 7.8 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.1 | 1.2 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.1 | 2.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 1.8 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 10.7 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.1 | 4.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 2.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.8 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 4.8 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 2.8 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.1 | 0.5 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 9.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 2.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 1.9 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 1.4 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 1.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.2 | GO:0016233 | telomere capping(GO:0016233) |
| 0.1 | 0.4 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 1.9 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 5.8 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 0.8 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.7 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.7 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.8 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 3.6 | GO:0048841 | regulation of axon extension involved in axon guidance(GO:0048841) |
| 0.1 | 0.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 5.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 6.5 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 2.8 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 1.0 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 1.6 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 3.7 | GO:0051051 | negative regulation of transport(GO:0051051) |
| 0.0 | 13.5 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.7 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 2.0 | GO:0090174 | vesicle fusion(GO:0006906) organelle membrane fusion(GO:0090174) |
| 0.0 | 1.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 10.2 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.9 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.5 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 2.8 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.9 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 3.5 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 0.3 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.1 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 6.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.5 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 2.7 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 6.8 | GO:0098916 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 2.1 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
| 0.0 | 0.6 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 4.9 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.8 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.8 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 6.9 | GO:0031175 | neuron projection development(GO:0031175) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 1.1 | 5.6 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.9 | 15.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.7 | 6.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.7 | 2.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.6 | 4.8 | GO:0035060 | brahma complex(GO:0035060) |
| 0.6 | 4.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.6 | 11.1 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.5 | 3.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.4 | 6.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.4 | 3.4 | GO:0071914 | prominosome(GO:0071914) |
| 0.4 | 1.2 | GO:1990879 | CST complex(GO:1990879) |
| 0.4 | 3.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.3 | 2.0 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.3 | 3.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.3 | 1.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 41.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.3 | 4.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.3 | 3.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.3 | 2.9 | GO:0070449 | elongin complex(GO:0070449) |
| 0.3 | 6.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 7.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 4.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.2 | 5.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 2.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.2 | 1.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 1.4 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.2 | 7.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 11.9 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 1.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 1.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 2.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.4 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.1 | 0.8 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 2.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 26.4 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 8.3 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 1.0 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 1.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 9.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 4.4 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 2.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 21.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 2.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.8 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 4.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.9 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 6.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.8 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.1 | 5.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 13.4 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.1 | 3.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 7.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 19.8 | GO:0043005 | neuron projection(GO:0043005) |
| 0.1 | 3.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 1.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 14.1 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 4.7 | GO:0098858 | actin-based cell projection(GO:0098858) |
| 0.1 | 4.6 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
| 0.0 | 0.6 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.0 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.4 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 3.1 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 1.2 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 0.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.7 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 1.1 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 15.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 25.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 4.8 | 19.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 2.8 | 8.5 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 2.6 | 7.9 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 2.0 | 6.0 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 1.9 | 5.6 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 1.3 | 5.2 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 1.3 | 5.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 1.2 | 3.6 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 1.1 | 7.8 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.9 | 5.6 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.9 | 4.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.8 | 12.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.8 | 10.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.8 | 3.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.8 | 5.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.8 | 5.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.6 | 8.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.5 | 9.9 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.5 | 2.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.5 | 2.5 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.5 | 6.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.5 | 10.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.5 | 3.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.4 | 2.5 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.4 | 4.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.4 | 16.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.4 | 0.8 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) type 1 melanocortin receptor binding(GO:0070996) |
| 0.4 | 2.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.4 | 3.0 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.4 | 9.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.4 | 2.8 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.4 | 4.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.4 | 4.6 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.3 | 2.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.3 | 1.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.3 | 6.0 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.3 | 1.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.2 | 1.5 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.2 | 8.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.2 | 2.7 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.2 | 9.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 6.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 1.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.2 | 9.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 3.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.3 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 2.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.4 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.1 | 3.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.4 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 2.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 1.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 2.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.2 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.1 | 1.0 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.1 | 1.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.8 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 1.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 6.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 8.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.7 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 4.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 9.9 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 3.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 2.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 2.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.8 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 26.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 12.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 3.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.9 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 3.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.7 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 1.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 2.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 2.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.7 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 1.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.7 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 1.5 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 1.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.8 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 8.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 2.1 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 3.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 3.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.8 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 1.6 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 3.0 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 3.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.4 | 4.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 18.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 4.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 1.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 1.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 2.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 5.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 5.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 6.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 5.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 11.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 1.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 3.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.9 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 19.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.6 | 5.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.6 | 8.3 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.5 | 4.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.5 | 5.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.4 | 4.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.4 | 1.5 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.4 | 5.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.4 | 7.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.3 | 5.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.3 | 1.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.3 | 5.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 1.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 2.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 8.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 5.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 3.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 4.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 2.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 2.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 3.0 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 1.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 1.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 4.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 2.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.4 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |