Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
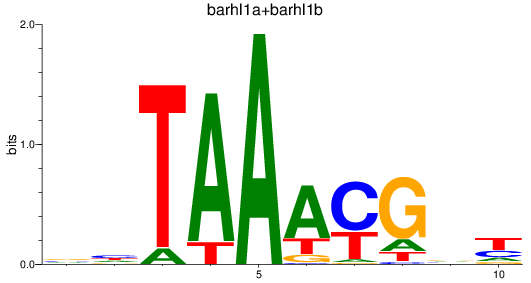
Results for barhl1a+barhl1b
Z-value: 1.70
Transcription factors associated with barhl1a+barhl1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
barhl1b
|
ENSDARG00000019013 | BarH-like homeobox 1b |
|
barhl1a
|
ENSDARG00000035508 | BarH-like homeobox 1a |
|
barhl1a
|
ENSDARG00000110061 | BarH-like homeobox 1a |
|
barhl1a
|
ENSDARG00000112355 | BarH-like homeobox 1a |
|
barhl1b
|
ENSDARG00000113145 | BarH-like homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| barhl1a | dr11_v1_chr5_-_29750377_29750377 | 0.57 | 1.6e-09 | Click! |
| barhl1b | dr11_v1_chr21_-_17482465_17482465 | 0.48 | 9.5e-07 | Click! |
Activity profile of barhl1a+barhl1b motif
Sorted Z-values of barhl1a+barhl1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_34701848 | 14.44 |
ENSDART00000082066
|
atpv0e2
|
ATPase H+ transporting V0 subunit e2 |
| chr14_-_1990290 | 11.60 |
ENSDART00000183382
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr8_+_25761654 | 10.54 |
ENSDART00000137899
ENSDART00000062403 |
tmem9
|
transmembrane protein 9 |
| chr2_-_32387441 | 10.40 |
ENSDART00000148202
|
ubtfl
|
upstream binding transcription factor, like |
| chr5_-_22082918 | 10.21 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr20_-_34801181 | 8.85 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr20_-_29475172 | 8.76 |
ENSDART00000183164
|
scg5
|
secretogranin V |
| chr3_+_35005730 | 8.74 |
ENSDART00000029451
|
prkcbb
|
protein kinase C, beta b |
| chr3_+_40170216 | 8.37 |
ENSDART00000011568
|
syngr3a
|
synaptogyrin 3a |
| chr2_-_30460293 | 7.97 |
ENSDART00000113193
|
cbln2a
|
cerebellin 2a precursor |
| chr2_-_13216269 | 7.11 |
ENSDART00000149947
|
bcl2b
|
BCL2, apoptosis regulator b |
| chr23_+_35713557 | 6.83 |
ENSDART00000123518
|
tuba1c
|
tubulin, alpha 1c |
| chr13_-_9525527 | 6.78 |
ENSDART00000190618
|
CR848040.5
|
|
| chr10_-_8033468 | 6.73 |
ENSDART00000140476
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr6_+_23752593 | 6.61 |
ENSDART00000164366
|
zgc:158654
|
zgc:158654 |
| chr14_-_2202652 | 6.56 |
ENSDART00000171316
|
si:dkey-262j3.7
|
si:dkey-262j3.7 |
| chr25_-_20666328 | 6.52 |
ENSDART00000098076
|
csk
|
C-terminal Src kinase |
| chr24_-_22756508 | 6.51 |
ENSDART00000035409
ENSDART00000146247 |
zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr6_-_21873266 | 6.43 |
ENSDART00000151658
ENSDART00000151152 ENSDART00000151179 |
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr18_+_31410652 | 6.40 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr22_+_10215558 | 6.40 |
ENSDART00000063274
|
kctd6a
|
potassium channel tetramerization domain containing 6a |
| chr23_-_24682244 | 6.30 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr20_+_33294428 | 6.23 |
ENSDART00000024104
|
mycn
|
MYCN proto-oncogene, bHLH transcription factor |
| chr5_+_32141790 | 6.15 |
ENSDART00000041504
|
tescb
|
tescalcin b |
| chr19_-_10394931 | 6.11 |
ENSDART00000191549
|
zgc:194578
|
zgc:194578 |
| chr18_+_7639401 | 6.06 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr14_+_50937757 | 6.05 |
ENSDART00000163865
|
rnf44
|
ring finger protein 44 |
| chr15_+_32249062 | 6.05 |
ENSDART00000133867
ENSDART00000152545 ENSDART00000082315 ENSDART00000152513 ENSDART00000152139 |
arfip2a
|
ADP-ribosylation factor interacting protein 2a |
| chr10_-_8032885 | 6.00 |
ENSDART00000188619
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr17_+_24446353 | 5.96 |
ENSDART00000140467
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr18_-_14743659 | 5.92 |
ENSDART00000125979
|
tshz3a
|
teashirt zinc finger homeobox 3a |
| chr3_-_20106233 | 5.86 |
ENSDART00000145285
|
selenow2b
|
selenoprotein W, 2b |
| chr5_-_28915130 | 5.75 |
ENSDART00000078592
|
npdc1b
|
neural proliferation, differentiation and control, 1b |
| chr3_+_45365098 | 5.66 |
ENSDART00000052746
ENSDART00000156555 |
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr3_-_19368435 | 5.64 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr19_+_342094 | 5.61 |
ENSDART00000151013
ENSDART00000187622 |
ensaa
|
endosulfine alpha a |
| chr3_+_45364849 | 5.46 |
ENSDART00000153974
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr17_+_24445818 | 5.41 |
ENSDART00000139963
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr7_+_25000060 | 5.36 |
ENSDART00000039265
ENSDART00000141814 |
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr17_-_33716688 | 5.31 |
ENSDART00000043651
|
dnal1
|
dynein, axonemal, light chain 1 |
| chr20_-_30938184 | 5.28 |
ENSDART00000147359
ENSDART00000062552 |
wtap
|
WT1 associated protein |
| chr10_+_21793670 | 5.14 |
ENSDART00000168918
|
pcdh1gc6
|
protocadherin 1 gamma c 6 |
| chr7_+_40228422 | 5.12 |
ENSDART00000052222
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr17_+_24446533 | 5.09 |
ENSDART00000131417
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr2_+_16449627 | 5.02 |
ENSDART00000005381
|
zgc:110269
|
zgc:110269 |
| chr2_-_37140423 | 4.99 |
ENSDART00000144220
|
tspan37
|
tetraspanin 37 |
| chr10_-_11397590 | 4.99 |
ENSDART00000064212
|
srek1ip1
|
SREK1-interacting protein 1 |
| chr16_-_31824525 | 4.98 |
ENSDART00000058737
|
cdc42l
|
cell division cycle 42, like |
| chr7_-_31921687 | 4.94 |
ENSDART00000146720
|
lin7c
|
lin-7 homolog C (C. elegans) |
| chr12_-_13318944 | 4.94 |
ENSDART00000152201
ENSDART00000041394 |
emc9
|
ER membrane protein complex subunit 9 |
| chr14_-_14705335 | 4.87 |
ENSDART00000157392
ENSDART00000159456 |
ogt.1
|
O-linked N-acetylglucosamine (GlcNAc) transferase, tandem duplicate 1 |
| chr13_-_28263856 | 4.87 |
ENSDART00000041036
ENSDART00000079806 |
smap1
|
small ArfGAP 1 |
| chr19_+_29798064 | 4.86 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
| chr13_+_46718518 | 4.85 |
ENSDART00000160401
ENSDART00000182884 |
tmem63ba
|
transmembrane protein 63Ba |
| chr19_-_33274978 | 4.82 |
ENSDART00000020301
ENSDART00000114714 |
fam92a1
|
family with sequence similarity 92, member A1 |
| chr3_-_52683241 | 4.82 |
ENSDART00000155248
|
si:dkey-210j14.5
|
si:dkey-210j14.5 |
| chr5_-_69437422 | 4.79 |
ENSDART00000073676
|
isca1
|
iron-sulfur cluster assembly 1 |
| chr19_+_46237665 | 4.78 |
ENSDART00000159391
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr6_-_55309190 | 4.78 |
ENSDART00000162117
|
ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr25_-_11088839 | 4.78 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr23_+_19813677 | 4.76 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr3_-_27868183 | 4.72 |
ENSDART00000185812
|
abat
|
4-aminobutyrate aminotransferase |
| chr8_-_10949847 | 4.71 |
ENSDART00000123209
|
pqlc2
|
PQ loop repeat containing 2 |
| chr22_+_11756040 | 4.56 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr7_+_24645186 | 4.55 |
ENSDART00000150118
ENSDART00000150233 ENSDART00000087691 |
gba2
|
glucosidase, beta (bile acid) 2 |
| chr3_+_31093455 | 4.52 |
ENSDART00000153074
|
si:dkey-66i24.9
|
si:dkey-66i24.9 |
| chr23_-_25779995 | 4.51 |
ENSDART00000110670
|
si:dkey-21c19.3
|
si:dkey-21c19.3 |
| chr22_-_9896180 | 4.44 |
ENSDART00000138343
|
znf990
|
zinc finger protein 990 |
| chr19_-_13950231 | 4.43 |
ENSDART00000168665
|
sh3bgrl3
|
SH3 domain binding glutamate-rich protein like 3 |
| chr23_+_27051919 | 4.41 |
ENSDART00000054237
|
ptges3a
|
prostaglandin E synthase 3a (cytosolic) |
| chr4_-_20051141 | 4.39 |
ENSDART00000066963
|
atp6v1f
|
ATPase H+ transporting V1 subunit F |
| chr19_-_21832441 | 4.34 |
ENSDART00000151272
ENSDART00000151442 ENSDART00000150168 ENSDART00000148797 ENSDART00000128196 ENSDART00000149259 ENSDART00000052556 ENSDART00000149658 ENSDART00000149639 ENSDART00000148424 |
mbpa
|
myelin basic protein a |
| chr25_-_31118923 | 4.29 |
ENSDART00000009126
ENSDART00000188286 |
kras
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
| chr15_-_19128705 | 4.28 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr19_-_3488860 | 4.25 |
ENSDART00000172520
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr12_-_27031060 | 4.24 |
ENSDART00000076103
|
chmp2a
|
charged multivesicular body protein 2A |
| chr22_-_28079678 | 4.20 |
ENSDART00000169887
|
wu:fu71h07
|
wu:fu71h07 |
| chr8_-_44223473 | 4.15 |
ENSDART00000098525
|
stx2b
|
syntaxin 2b |
| chr2_-_26469065 | 4.09 |
ENSDART00000099247
ENSDART00000099248 |
rabggtb
|
Rab geranylgeranyltransferase, beta subunit |
| chr2_-_13268556 | 4.08 |
ENSDART00000127693
ENSDART00000079757 |
vps4b
|
vacuolar protein sorting 4b homolog B (S. cerevisiae) |
| chr1_+_34696503 | 4.05 |
ENSDART00000186106
|
CR339054.2
|
|
| chr2_-_32826108 | 4.04 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr25_-_26753196 | 4.02 |
ENSDART00000155698
|
usp3
|
ubiquitin specific peptidase 3 |
| chr17_+_22311413 | 3.98 |
ENSDART00000151929
|
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr22_-_10586191 | 3.97 |
ENSDART00000148418
|
si:dkey-42i9.16
|
si:dkey-42i9.16 |
| chr22_-_33916620 | 3.96 |
ENSDART00000191276
|
CR974456.1
|
|
| chr23_+_39854566 | 3.93 |
ENSDART00000190423
ENSDART00000164473 ENSDART00000161881 |
si:ch73-217b7.1
|
si:ch73-217b7.1 |
| chr8_-_44223899 | 3.92 |
ENSDART00000143020
|
stx2b
|
syntaxin 2b |
| chr3_-_18274691 | 3.89 |
ENSDART00000161140
|
BX649434.3
|
|
| chr19_+_29808471 | 3.88 |
ENSDART00000186428
|
hdac1
|
histone deacetylase 1 |
| chr7_+_13418812 | 3.87 |
ENSDART00000191905
ENSDART00000091567 |
dagla
|
diacylglycerol lipase, alpha |
| chr13_-_24379271 | 3.86 |
ENSDART00000046360
|
rhoua
|
ras homolog family member Ua |
| chr23_-_12015139 | 3.85 |
ENSDART00000110627
ENSDART00000193988 ENSDART00000184528 |
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr4_+_20051478 | 3.82 |
ENSDART00000143642
|
lamtor4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr2_-_31614277 | 3.80 |
ENSDART00000137054
ENSDART00000131507 ENSDART00000137674 ENSDART00000147041 |
si:ch211-106h4.5
|
si:ch211-106h4.5 |
| chr1_+_8601935 | 3.72 |
ENSDART00000152367
|
si:ch211-160d14.6
|
si:ch211-160d14.6 |
| chr20_-_32110882 | 3.72 |
ENSDART00000030324
|
grm1a
|
glutamate receptor, metabotropic 1a |
| chr20_-_10120442 | 3.70 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr7_+_26466826 | 3.69 |
ENSDART00000058908
|
mpdu1b
|
mannose-P-dolichol utilization defect 1b |
| chr19_+_18739085 | 3.68 |
ENSDART00000188868
|
spaca4l
|
sperm acrosome associated 4 like |
| chr15_-_19669770 | 3.68 |
ENSDART00000152707
|
si:dkey-4p15.3
|
si:dkey-4p15.3 |
| chr2_-_32825917 | 3.61 |
ENSDART00000180409
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr13_+_15656042 | 3.58 |
ENSDART00000134240
|
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr4_+_30363474 | 3.51 |
ENSDART00000168421
|
si:dkey-199m13.7
|
si:dkey-199m13.7 |
| chr3_-_61387273 | 3.46 |
ENSDART00000156479
|
znf1143
|
zinc finger protein 1143 |
| chr3_-_15352303 | 3.44 |
ENSDART00000104338
ENSDART00000145919 ENSDART00000132135 |
pitpnbl
|
phosphatidylinositol transfer protein, beta, like |
| chr9_-_33477588 | 3.43 |
ENSDART00000144150
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr3_-_32859335 | 3.41 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr25_+_17590095 | 3.41 |
ENSDART00000009767
|
vac14
|
vac14 homolog (S. cerevisiae) |
| chr7_+_34236238 | 3.40 |
ENSDART00000052474
|
tipin
|
timeless interacting protein |
| chr11_+_6136220 | 3.39 |
ENSDART00000082223
|
tax1bp3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr18_+_7150144 | 3.38 |
ENSDART00000114690
|
vwf
|
von Willebrand factor |
| chr17_-_14451718 | 3.37 |
ENSDART00000039438
|
jkamp
|
jnk1/mapk8 associated membrane protein |
| chr21_-_2162850 | 3.37 |
ENSDART00000159731
|
gb:ai877918
|
expressed sequence AI877918 |
| chr20_-_32148901 | 3.35 |
ENSDART00000153405
ENSDART00000048537 ENSDART00000152984 |
cep57l1
|
centrosomal protein 57, like 1 |
| chr25_-_20268027 | 3.34 |
ENSDART00000138763
|
dnajb9a
|
DnaJ (Hsp40) homolog, subfamily B, member 9a |
| chr3_+_54761569 | 3.32 |
ENSDART00000135913
ENSDART00000180983 |
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr8_+_25190023 | 3.31 |
ENSDART00000047528
|
gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr3_-_52644737 | 3.30 |
ENSDART00000126180
|
si:dkey-210j14.3
|
si:dkey-210j14.3 |
| chr16_+_43347966 | 3.28 |
ENSDART00000171308
|
zmp:0000000930
|
zmp:0000000930 |
| chr24_-_41797681 | 3.28 |
ENSDART00000169643
|
arhgap28
|
Rho GTPase activating protein 28 |
| chr4_+_19535946 | 3.28 |
ENSDART00000192342
ENSDART00000183740 ENSDART00000180812 ENSDART00000180017 |
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr15_-_6976851 | 3.25 |
ENSDART00000158474
ENSDART00000168943 ENSDART00000169944 |
si:ch73-311h14.2
|
si:ch73-311h14.2 |
| chr2_-_38080075 | 3.24 |
ENSDART00000056544
|
tox4a
|
TOX high mobility group box family member 4 a |
| chr25_-_24202576 | 3.23 |
ENSDART00000048507
|
uevld
|
UEV and lactate/malate dehyrogenase domains |
| chr2_-_10338759 | 3.22 |
ENSDART00000150166
ENSDART00000149584 |
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr6_+_296130 | 3.20 |
ENSDART00000073985
|
rbfox2
|
RNA binding fox-1 homolog 2 |
| chr13_+_22717939 | 3.18 |
ENSDART00000188288
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr3_-_32590164 | 3.16 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr3_+_32714157 | 3.16 |
ENSDART00000131774
|
setd1a
|
SET domain containing 1A |
| chr3_+_22442445 | 3.14 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr1_+_29267841 | 3.12 |
ENSDART00000085648
|
lig4
|
ligase IV, DNA, ATP-dependent |
| chr16_-_35415004 | 3.12 |
ENSDART00000170401
ENSDART00000157832 ENSDART00000170048 |
taf12
|
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr11_-_33618612 | 3.11 |
ENSDART00000033980
|
lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr3_-_16110100 | 3.09 |
ENSDART00000051807
|
lasp1
|
LIM and SH3 protein 1 |
| chr3_-_36210344 | 3.08 |
ENSDART00000025326
|
csnk1da
|
casein kinase 1, delta a |
| chr22_-_21314821 | 3.07 |
ENSDART00000105546
ENSDART00000135388 |
cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr13_+_9612395 | 3.06 |
ENSDART00000136689
ENSDART00000138362 |
slf2
|
SMC5-SMC6 complex localization factor 2 |
| chr1_+_52481332 | 3.05 |
ENSDART00000074231
|
cldnd1b
|
claudin domain containing 1b |
| chr16_+_9400661 | 3.04 |
ENSDART00000146174
|
ice1
|
KIAA0947-like (H. sapiens) |
| chr5_-_35252761 | 3.04 |
ENSDART00000051278
|
tnpo1
|
transportin 1 |
| chr17_+_24597001 | 3.02 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr9_-_34300707 | 3.02 |
ENSDART00000049805
|
ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr5_-_32890807 | 3.02 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr12_-_7655322 | 2.97 |
ENSDART00000181162
|
ank3b
|
ankyrin 3b |
| chr19_+_14921000 | 2.97 |
ENSDART00000144052
|
oprd1a
|
opioid receptor, delta 1a |
| chr16_+_49601838 | 2.96 |
ENSDART00000168570
ENSDART00000159236 |
si:dkey-82o10.4
|
si:dkey-82o10.4 |
| chr4_-_75172216 | 2.94 |
ENSDART00000127522
|
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr3_+_13842554 | 2.93 |
ENSDART00000162317
ENSDART00000158068 |
ilf3b
|
interleukin enhancer binding factor 3b |
| chr5_+_57480014 | 2.90 |
ENSDART00000135520
|
si:ch211-202f5.3
|
si:ch211-202f5.3 |
| chr23_+_28582865 | 2.88 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr16_+_25407021 | 2.86 |
ENSDART00000187489
ENSDART00000086333 |
jarid2a
|
jumonji, AT rich interactive domain 2a |
| chr23_+_42272588 | 2.85 |
ENSDART00000164907
|
CABZ01065131.1
|
|
| chr21_-_25801956 | 2.84 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr3_+_26019426 | 2.83 |
ENSDART00000135389
ENSDART00000182411 |
foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr18_+_17663898 | 2.82 |
ENSDART00000021213
|
cpne2
|
copine II |
| chr8_-_7847921 | 2.81 |
ENSDART00000188094
|
zgc:113363
|
zgc:113363 |
| chr12_+_16087077 | 2.81 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr9_+_23748342 | 2.80 |
ENSDART00000019053
|
faima
|
Fas apoptotic inhibitory molecule a |
| chr11_+_37612214 | 2.79 |
ENSDART00000172899
ENSDART00000077496 |
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr12_+_28910762 | 2.77 |
ENSDART00000076342
ENSDART00000160939 ENSDART00000076572 |
rnf40
|
ring finger protein 40 |
| chr24_-_6345647 | 2.77 |
ENSDART00000108994
|
zgc:174877
|
zgc:174877 |
| chr13_-_11072964 | 2.73 |
ENSDART00000135989
|
cep170aa
|
centrosomal protein 170Aa |
| chr14_+_8947282 | 2.73 |
ENSDART00000047993
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr12_-_20409794 | 2.73 |
ENSDART00000077936
|
lcmt1
|
leucine carboxyl methyltransferase 1 |
| chr5_+_53580846 | 2.72 |
ENSDART00000184967
ENSDART00000161751 |
taf6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr19_+_31904836 | 2.72 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr24_-_11057305 | 2.72 |
ENSDART00000186494
|
asap1b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1b |
| chr6_+_9107063 | 2.71 |
ENSDART00000083820
|
vps16
|
vacuolar protein sorting protein 16 |
| chr17_+_21760032 | 2.71 |
ENSDART00000190425
|
ikzf5
|
IKAROS family zinc finger 5 |
| chr17_+_21817382 | 2.70 |
ENSDART00000079011
ENSDART00000189387 |
ikzf5
|
IKAROS family zinc finger 5 |
| chr5_+_57714902 | 2.69 |
ENSDART00000182860
|
ufc1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chr3_+_27798094 | 2.69 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr1_+_19433004 | 2.69 |
ENSDART00000133959
|
clockb
|
clock circadian regulator b |
| chr4_+_15006217 | 2.69 |
ENSDART00000090837
|
zc3hc1
|
zinc finger, C3HC-type containing 1 |
| chr5_-_42180205 | 2.67 |
ENSDART00000145247
|
fam222ba
|
family with sequence similarity 222, member Ba |
| chr2_+_15776156 | 2.63 |
ENSDART00000190795
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr12_+_18906939 | 2.59 |
ENSDART00000186074
|
josd1
|
Josephin domain containing 1 |
| chr1_-_31515746 | 2.59 |
ENSDART00000190886
|
cenpk
|
centromere protein K |
| chr7_-_40630698 | 2.59 |
ENSDART00000134547
|
ube3c
|
ubiquitin protein ligase E3C |
| chr2_+_32826235 | 2.58 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr16_-_11779508 | 2.58 |
ENSDART00000136329
ENSDART00000060145 ENSDART00000141101 |
pafah1b3
|
platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit |
| chr7_+_21887307 | 2.56 |
ENSDART00000052871
|
pop7
|
POP7 homolog, ribonuclease P/MRP subunit |
| chr5_+_36900157 | 2.55 |
ENSDART00000183533
ENSDART00000051184 |
hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr23_+_24955394 | 2.54 |
ENSDART00000142124
|
nol9
|
nucleolar protein 9 |
| chr13_+_9368621 | 2.53 |
ENSDART00000109126
|
alms1
|
Alstrom syndrome protein 1 |
| chr14_+_21820034 | 2.51 |
ENSDART00000122739
|
ctbp1
|
C-terminal binding protein 1 |
| chr1_+_26105141 | 2.49 |
ENSDART00000102379
ENSDART00000127154 |
toporsa
|
topoisomerase I binding, arginine/serine-rich a |
| chr24_+_12075009 | 2.48 |
ENSDART00000181141
|
ccr9b
|
chemokine (C-C motif) receptor 9b |
| chr21_-_30181732 | 2.47 |
ENSDART00000015636
|
hnrnph1l
|
heterogeneous nuclear ribonucleoprotein H1, like |
| chr12_+_18681477 | 2.47 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr14_-_2206476 | 2.46 |
ENSDART00000081870
|
pcdh2ab6
|
protocadherin 2 alpha b 6 |
| chr8_-_16674584 | 2.43 |
ENSDART00000100727
|
osbpl9
|
oxysterol binding protein-like 9 |
| chr7_+_24081167 | 2.43 |
ENSDART00000141749
|
acin1b
|
apoptotic chromatin condensation inducer 1b |
| chr7_-_7972445 | 2.43 |
ENSDART00000182687
ENSDART00000191946 |
si:cabz01030277.1
|
si:cabz01030277.1 |
| chr12_+_5209822 | 2.42 |
ENSDART00000152610
|
SLC35G1
|
si:ch211-197g18.2 |
| chr3_+_7040363 | 2.41 |
ENSDART00000157805
|
BX000701.2
|
|
| chr22_-_10055744 | 2.41 |
ENSDART00000143686
|
si:ch211-222k6.2
|
si:ch211-222k6.2 |
| chr10_-_42108137 | 2.40 |
ENSDART00000132976
|
ess2
|
ess-2 splicing factor homolog |
| chr19_-_20777351 | 2.40 |
ENSDART00000019206
|
ngly1
|
N-glycanase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of barhl1a+barhl1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 16.5 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 2.8 | 11.1 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 1.5 | 4.6 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 1.3 | 6.5 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 1.2 | 4.7 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 1.1 | 5.7 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 1.1 | 3.4 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 1.0 | 3.0 | GO:0006272 | leading strand elongation(GO:0006272) |
| 1.0 | 4.9 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 1.0 | 8.8 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 1.0 | 5.9 | GO:0010269 | response to selenium ion(GO:0010269) |
| 1.0 | 3.9 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.9 | 4.7 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.9 | 6.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.9 | 12.7 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.9 | 4.5 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.9 | 5.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.8 | 4.0 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.8 | 2.3 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.8 | 3.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.7 | 3.0 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.7 | 2.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.7 | 2.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.7 | 4.8 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.7 | 3.4 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.6 | 8.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.6 | 3.0 | GO:0051204 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.6 | 4.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.6 | 2.9 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.6 | 3.4 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.6 | 3.9 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.5 | 4.8 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.5 | 3.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.5 | 2.5 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.5 | 1.9 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.5 | 2.4 | GO:0072672 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.4 | 1.8 | GO:0021543 | substrate-dependent cell migration(GO:0006929) pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 0.4 | 2.2 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.4 | 3.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.4 | 6.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.4 | 5.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.4 | 4.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.3 | 10.0 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.3 | 1.0 | GO:0000423 | macromitophagy(GO:0000423) |
| 0.3 | 1.0 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.3 | 2.7 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.3 | 4.4 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) chaperone-mediated protein complex assembly(GO:0051131) |
| 0.3 | 4.9 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.3 | 1.4 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.3 | 0.8 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.3 | 6.0 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.3 | 2.8 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.3 | 1.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.3 | 0.8 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.2 | 4.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.2 | 1.2 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.2 | 3.7 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.2 | 2.7 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.2 | 8.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.2 | 8.1 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.2 | 3.1 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.2 | 2.4 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.2 | 15.6 | GO:0044839 | cell cycle G2/M phase transition(GO:0044839) |
| 0.2 | 0.7 | GO:2000637 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.2 | 2.6 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.2 | 14.4 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.2 | 8.8 | GO:0046883 | regulation of hormone secretion(GO:0046883) |
| 0.2 | 2.3 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.2 | 2.3 | GO:0045453 | bone resorption(GO:0045453) |
| 0.2 | 2.0 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.2 | 4.0 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.2 | 2.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 | 3.3 | GO:0051098 | regulation of binding(GO:0051098) |
| 0.2 | 2.4 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 1.4 | GO:0086002 | cardiac muscle cell action potential involved in contraction(GO:0086002) |
| 0.2 | 1.7 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 1.7 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.2 | 5.3 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.2 | 2.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.2 | 3.8 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) cellular response to amino acid stimulus(GO:0071230) |
| 0.2 | 8.7 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 2.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 4.9 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 2.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 2.9 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 2.7 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 1.7 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 3.7 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 1.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 2.0 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 3.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 4.1 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.1 | 0.7 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.1 | 0.4 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 0.1 | 2.0 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 27.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.7 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 3.9 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.1 | 0.3 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 3.2 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 0.6 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 0.7 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.6 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 11.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 0.8 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.5 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 2.2 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 1.6 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.1 | 2.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 2.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 1.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.5 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.1 | 1.0 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.2 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.1 | 2.2 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.1 | 1.3 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 1.1 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.1 | 0.5 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 3.9 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 3.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 3.3 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.1 | 8.8 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 2.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 2.7 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.1 | 6.4 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 2.1 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 1.9 | GO:1901215 | negative regulation of neuron death(GO:1901215) |
| 0.0 | 0.3 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 2.8 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.9 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 2.7 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 4.9 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 1.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.7 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 2.9 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 1.9 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 1.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.2 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 2.0 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.5 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 2.1 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 10.3 | GO:0007188 | adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.0 | 0.8 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.2 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 1.3 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 4.4 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 1.7 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 2.6 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 2.1 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 1.9 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 1.2 | GO:0035194 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.0 | 2.3 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 2.2 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 5.8 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 1.3 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 2.2 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.4 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 2.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.9 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 3.4 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 0.2 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 1.5 | GO:0099518 | vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 1.6 | GO:0046939 | nucleotide phosphorylation(GO:0046939) |
| 0.0 | 1.7 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 7.1 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 0.5 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 2.3 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 1.1 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.0 | 1.0 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 1.5 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 1.3 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 2.4 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 2.8 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 1.8 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 2.6 | GO:0016042 | lipid catabolic process(GO:0016042) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 27.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 1.5 | 5.8 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 1.3 | 5.3 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 1.2 | 4.9 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 1.1 | 3.4 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 1.0 | 3.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 1.0 | 4.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.8 | 3.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.8 | 3.0 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.7 | 3.4 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.6 | 3.8 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.6 | 1.9 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.6 | 4.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.5 | 2.7 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.5 | 2.6 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.4 | 4.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.4 | 1.8 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.4 | 2.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.4 | 3.4 | GO:0035032 | extrinsic component of vacuolar membrane(GO:0000306) phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.3 | 1.7 | GO:0033503 | HULC complex(GO:0033503) |
| 0.3 | 8.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.3 | 1.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.3 | 4.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.3 | 4.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.3 | 6.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.3 | 4.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.3 | 1.7 | GO:0070695 | FHF complex(GO:0070695) |
| 0.3 | 8.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.3 | 1.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.3 | 2.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.3 | 4.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 2.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 3.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 4.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 0.9 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.2 | 3.8 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 2.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 1.5 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 1.5 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.2 | 15.7 | GO:0030141 | secretory granule(GO:0030141) |
| 0.2 | 2.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 6.1 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 6.8 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 1.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 5.0 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 1.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 3.7 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 1.0 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 2.1 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 6.8 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 4.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 3.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 3.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 7.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 0.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) contractile ring(GO:0070938) |
| 0.1 | 3.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 3.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 3.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 5.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 4.0 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 0.7 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.6 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 2.0 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 2.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 4.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 11.1 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 6.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 3.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 7.6 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 2.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.6 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.6 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.2 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 2.7 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 11.1 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 1.2 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 1.3 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 73.5 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 14.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.7 | GO:0070161 | adherens junction(GO:0005912) anchoring junction(GO:0070161) |
| 0.0 | 2.8 | GO:0005768 | endosome(GO:0005768) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 16.5 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 2.8 | 11.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 2.1 | 8.3 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 1.3 | 3.9 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 1.2 | 8.7 | GO:0035173 | histone kinase activity(GO:0035173) |
| 1.2 | 4.9 | GO:0097016 | L27 domain binding(GO:0097016) |
| 1.1 | 4.4 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 1.0 | 4.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 1.0 | 3.0 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.9 | 6.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.9 | 2.7 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.9 | 6.2 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.8 | 12.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.8 | 4.9 | GO:0097363 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.8 | 2.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.7 | 5.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.7 | 5.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.7 | 6.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.6 | 2.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.5 | 3.7 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.5 | 3.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.5 | 3.7 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.4 | 5.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.4 | 1.2 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.4 | 2.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.4 | 4.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.4 | 2.0 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.4 | 6.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.4 | 5.6 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.4 | 3.9 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.3 | 2.3 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 1.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 4.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.3 | 2.7 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.3 | 2.9 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 4.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 2.9 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.3 | 2.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.3 | 3.9 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 4.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 3.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.2 | 3.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 5.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 3.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 2.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 2.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 4.8 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.2 | 2.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 3.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 4.1 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.2 | 2.3 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.2 | 3.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 2.0 | GO:0022839 | ion gated channel activity(GO:0022839) |
| 0.2 | 2.9 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 3.0 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.2 | 1.9 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.2 | 10.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 1.7 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.2 | 1.0 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.2 | 1.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 4.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.2 | 1.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 1.7 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 0.6 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 4.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 7.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 4.3 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.1 | 0.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 2.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 3.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 6.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 2.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 2.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 4.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 3.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.5 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 0.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 7.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 6.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 11.5 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 0.3 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.1 | 1.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 7.1 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 1.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 2.2 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 1.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 7.2 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 2.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 1.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 2.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 7.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 17.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 3.4 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 4.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 5.7 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 3.8 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 5.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 2.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.7 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 32.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 9.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 10.0 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 2.8 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 1.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 3.7 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 5.4 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 5.3 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 3.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 5.4 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.3 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.8 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.5 | GO:0015103 | inorganic anion transmembrane transporter activity(GO:0015103) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.4 | 4.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.4 | 3.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 3.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 10.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.2 | 4.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 4.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 10.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 5.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 4.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.5 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 3.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 5.5 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 3.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 2.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.5 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.7 | 4.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.5 | 3.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.5 | 13.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.4 | 6.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.4 | 3.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.4 | 2.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.4 | 4.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.3 | 4.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 3.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 5.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.2 | 3.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.2 | 3.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 3.0 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.2 | 2.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 1.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 3.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 2.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 4.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 1.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 3.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.2 | 2.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 3.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 5.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 5.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 1.1 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 1.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 2.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 2.5 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 3.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 2.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |