Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
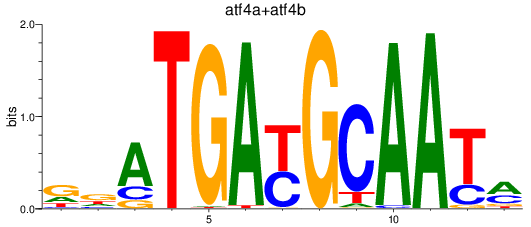
Results for atf4a+atf4b
Z-value: 1.34
Transcription factors associated with atf4a+atf4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
atf4b
|
ENSDARG00000038141 | activating transcription factor 4b |
|
atf4a
|
ENSDARG00000111939 | activating transcription factor 4a |
|
atf4b
|
ENSDARG00000114163 | activating transcription factor 4b |
|
atf4b
|
ENSDARG00000114542 | activating transcription factor 4b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| atf4b | dr11_v1_chr3_+_24197934_24197934 | 0.44 | 9.1e-06 | Click! |
| atf4a | dr11_v1_chr6_+_269204_269204 | -0.28 | 5.6e-03 | Click! |
Activity profile of atf4a+atf4b motif
Sorted Z-values of atf4a+atf4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_31119548 | 14.41 |
ENSDART00000136578
|
syn1
|
synapsin I |
| chr16_+_34523515 | 14.01 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr3_+_31933893 | 12.31 |
ENSDART00000146509
ENSDART00000139644 |
lin7b
|
lin-7 homolog B (C. elegans) |
| chr15_-_24869826 | 11.66 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr4_+_17279966 | 11.56 |
ENSDART00000067005
ENSDART00000137487 |
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr19_-_9712530 | 10.65 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr15_+_29085955 | 10.24 |
ENSDART00000156799
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr21_+_30502002 | 10.03 |
ENSDART00000043727
|
slc7a3a
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3a |
| chr4_+_1600034 | 9.90 |
ENSDART00000146779
|
slc38a2
|
solute carrier family 38, member 2 |
| chr5_-_55395964 | 9.90 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr4_+_8797197 | 9.81 |
ENSDART00000158671
|
sult4a1
|
sulfotransferase family 4A, member 1 |
| chr14_+_28438947 | 9.44 |
ENSDART00000006489
|
acsl4a
|
acyl-CoA synthetase long chain family member 4a |
| chr12_+_46960579 | 8.46 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr7_-_52842605 | 8.36 |
ENSDART00000083002
|
map1aa
|
microtubule-associated protein 1Aa |
| chr23_-_27345425 | 7.79 |
ENSDART00000022042
ENSDART00000191870 |
scn8aa
|
sodium channel, voltage gated, type VIII, alpha subunit a |
| chr9_-_9228941 | 7.67 |
ENSDART00000121665
|
cbsb
|
cystathionine-beta-synthase b |
| chr5_-_32323136 | 7.44 |
ENSDART00000110804
|
hspb15
|
heat shock protein, alpha-crystallin-related, b15 |
| chr6_+_41255485 | 7.39 |
ENSDART00000042683
ENSDART00000186013 |
cadpsb
|
Ca2+-dependent activator protein for secretion b |
| chr7_+_33314925 | 7.37 |
ENSDART00000148590
|
coro2ba
|
coronin, actin binding protein, 2Ba |
| chr17_-_45552602 | 7.18 |
ENSDART00000154844
ENSDART00000034432 |
susd4
|
sushi domain containing 4 |
| chr6_-_11780070 | 6.94 |
ENSDART00000151195
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr2_-_36932253 | 6.92 |
ENSDART00000124217
|
map1sb
|
microtubule-associated protein 1Sb |
| chr5_-_69499486 | 6.88 |
ENSDART00000023983
ENSDART00000180293 |
psat1
|
phosphoserine aminotransferase 1 |
| chr13_-_24447332 | 6.82 |
ENSDART00000043004
|
slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr18_-_16179129 | 6.50 |
ENSDART00000125353
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr25_+_7784582 | 6.32 |
ENSDART00000155016
|
dgkzb
|
diacylglycerol kinase, zeta b |
| chr19_+_30885258 | 6.27 |
ENSDART00000143394
|
yars
|
tyrosyl-tRNA synthetase |
| chr5_-_57528943 | 6.26 |
ENSDART00000130320
|
pisd
|
phosphatidylserine decarboxylase |
| chr12_+_34953038 | 5.98 |
ENSDART00000187022
ENSDART00000123988 ENSDART00000027034 |
qki2
|
QKI, KH domain containing, RNA binding 2 |
| chr21_-_43949208 | 5.79 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr18_+_17493859 | 5.60 |
ENSDART00000090754
|
si:dkey-102f14.5
|
si:dkey-102f14.5 |
| chr5_-_55395384 | 5.36 |
ENSDART00000147298
ENSDART00000082577 |
prune2
|
prune homolog 2 (Drosophila) |
| chr9_+_2574122 | 5.27 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr5_+_3501859 | 5.08 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr20_+_16743056 | 5.03 |
ENSDART00000050308
|
calm1b
|
calmodulin 1b |
| chr3_+_33440615 | 4.93 |
ENSDART00000146005
|
gtpbp1
|
GTP binding protein 1 |
| chr12_-_32066469 | 4.90 |
ENSDART00000140685
ENSDART00000062185 |
rab40b
|
RAB40B, member RAS oncogene family |
| chr2_+_35240764 | 4.82 |
ENSDART00000015827
|
tnr
|
tenascin R (restrictin, janusin) |
| chr9_-_48214216 | 4.75 |
ENSDART00000012938
|
phgdh
|
phosphoglycerate dehydrogenase |
| chr23_-_12014931 | 4.57 |
ENSDART00000134652
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr15_-_25392589 | 4.48 |
ENSDART00000124205
|
si:dkey-54n8.4
|
si:dkey-54n8.4 |
| chr19_-_30447611 | 4.33 |
ENSDART00000073705
ENSDART00000048977 ENSDART00000191237 |
abcf1
|
ATP-binding cassette, sub-family F (GCN20), member 1 |
| chr19_+_30884706 | 4.32 |
ENSDART00000052126
|
yars
|
tyrosyl-tRNA synthetase |
| chr20_-_20610812 | 4.16 |
ENSDART00000181870
|
ppm1ab
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ab |
| chr16_+_40024883 | 3.93 |
ENSDART00000110100
|
hint3
|
histidine triad nucleotide binding protein 3 |
| chr17_+_12700617 | 3.85 |
ENSDART00000191931
|
stmn4l
|
stathmin-like 4, like |
| chr12_+_41697664 | 3.85 |
ENSDART00000162302
|
bnip3
|
BCL2 interacting protein 3 |
| chr20_-_29418620 | 3.85 |
ENSDART00000172634
|
ryr3
|
ryanodine receptor 3 |
| chr20_-_20611063 | 3.79 |
ENSDART00000063492
|
ppm1ab
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ab |
| chr3_-_40284744 | 3.69 |
ENSDART00000018626
|
pdap1b
|
pdgfa associated protein 1b |
| chr20_-_35246150 | 3.53 |
ENSDART00000090549
|
fzd3a
|
frizzled class receptor 3a |
| chr3_-_30152836 | 3.52 |
ENSDART00000165920
|
nucb1
|
nucleobindin 1 |
| chr3_-_30153242 | 3.45 |
ENSDART00000077089
|
nucb1
|
nucleobindin 1 |
| chr12_-_46959990 | 3.44 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr20_-_19532531 | 3.41 |
ENSDART00000125830
ENSDART00000179566 ENSDART00000184193 |
eif2b4
BX276101.2
|
eukaryotic translation initiation factor 2B, subunit 4 delta |
| chr19_+_30884960 | 3.37 |
ENSDART00000140603
ENSDART00000183224 ENSDART00000135484 ENSDART00000139599 |
yars
|
tyrosyl-tRNA synthetase |
| chr22_+_25931782 | 3.33 |
ENSDART00000157842
|
dnaja3b
|
DnaJ (Hsp40) homolog, subfamily A, member 3B |
| chr6_+_49551614 | 3.32 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr7_-_31830936 | 3.31 |
ENSDART00000052514
ENSDART00000129720 |
cars
|
cysteinyl-tRNA synthetase |
| chr6_+_58915889 | 3.24 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr24_+_32525146 | 3.21 |
ENSDART00000132417
ENSDART00000110185 |
yme1l1a
|
YME1-like 1a |
| chr23_-_18030399 | 3.17 |
ENSDART00000136967
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr21_+_944715 | 3.10 |
ENSDART00000144766
|
nars
|
asparaginyl-tRNA synthetase |
| chr6_+_37754763 | 3.07 |
ENSDART00000110770
|
herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr6_-_37422841 | 3.07 |
ENSDART00000138351
|
cth
|
cystathionase (cystathionine gamma-lyase) |
| chr6_-_31233696 | 3.07 |
ENSDART00000079173
|
lepr
|
leptin receptor |
| chr12_-_35386910 | 3.06 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr8_+_42917515 | 3.01 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr6_+_23809501 | 3.01 |
ENSDART00000168701
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr13_-_25548733 | 2.96 |
ENSDART00000168099
ENSDART00000135788 ENSDART00000077655 |
mcmbp
|
minichromosome maintenance complex binding protein |
| chr10_-_31104983 | 2.94 |
ENSDART00000186941
|
pknox2
|
pbx/knotted 1 homeobox 2 |
| chr12_+_27129659 | 2.79 |
ENSDART00000076161
|
hoxb5b
|
homeobox B5b |
| chr12_-_13205572 | 2.78 |
ENSDART00000152670
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr12_+_30789611 | 2.77 |
ENSDART00000181501
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr16_-_20006270 | 2.75 |
ENSDART00000079155
|
hdac9b
|
histone deacetylase 9b |
| chr20_+_37794633 | 2.74 |
ENSDART00000022060
|
atf3
|
activating transcription factor 3 |
| chr7_-_35036770 | 2.64 |
ENSDART00000123174
|
galr1b
|
galanin receptor 1b |
| chr5_-_57652168 | 2.52 |
ENSDART00000166245
|
gb:eh456644
|
expressed sequence EH456644 |
| chr2_+_7849890 | 2.30 |
ENSDART00000114241
|
si:ch211-38m6.6
|
si:ch211-38m6.6 |
| chr25_+_28893615 | 2.28 |
ENSDART00000156994
ENSDART00000075151 |
amn1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr2_-_37059966 | 2.25 |
ENSDART00000137967
|
diras1b
|
DIRAS family, GTP-binding RAS-like 1b |
| chr13_+_4225173 | 2.22 |
ENSDART00000058242
ENSDART00000143456 |
mea1
|
male-enhanced antigen 1 |
| chr18_-_18667416 | 2.15 |
ENSDART00000100401
ENSDART00000136544 |
aars
|
alanyl-tRNA synthetase |
| chr12_-_48374728 | 2.12 |
ENSDART00000153403
ENSDART00000188117 |
dnajb12b
|
DnaJ (Hsp40) homolog, subfamily B, member 12b |
| chr15_-_23908779 | 2.12 |
ENSDART00000088808
|
usp32
|
ubiquitin specific peptidase 32 |
| chr15_-_2754056 | 2.12 |
ENSDART00000129380
|
ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr23_+_35504824 | 2.11 |
ENSDART00000082647
ENSDART00000159218 |
acp1
|
acid phosphatase 1 |
| chr12_+_30788912 | 2.06 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr7_-_30272871 | 2.05 |
ENSDART00000099586
|
zgc:162945
|
zgc:162945 |
| chr20_+_28266892 | 2.04 |
ENSDART00000103330
|
chac1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr8_-_46386024 | 2.03 |
ENSDART00000136602
ENSDART00000060919 ENSDART00000137472 |
qars
|
glutaminyl-tRNA synthetase |
| chr14_-_30945515 | 2.00 |
ENSDART00000161540
|
si:zfos-80g12.1
|
si:zfos-80g12.1 |
| chr2_+_1001560 | 1.93 |
ENSDART00000134656
|
cacna1eb
|
calcium channel, voltage-dependent, R type, alpha 1E subunit b |
| chr15_-_42283830 | 1.92 |
ENSDART00000015979
|
farsb
|
phenylalanyl-tRNA synthetase, beta subunit |
| chr12_-_13205854 | 1.91 |
ENSDART00000077829
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr2_-_28671139 | 1.87 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr3_+_15394428 | 1.86 |
ENSDART00000133168
|
atxn2l
|
ataxin 2-like |
| chr20_-_18915376 | 1.85 |
ENSDART00000063725
|
xkr6b
|
XK, Kell blood group complex subunit-related family, member 6b |
| chr3_+_25907266 | 1.81 |
ENSDART00000170324
ENSDART00000192633 |
tom1
|
target of myb1 membrane trafficking protein |
| chr15_-_17870090 | 1.80 |
ENSDART00000155066
|
atf5b
|
activating transcription factor 5b |
| chr2_+_16597011 | 1.79 |
ENSDART00000160641
ENSDART00000125413 |
xrn1
|
5'-3' exoribonuclease 1 |
| chr8_-_31384607 | 1.78 |
ENSDART00000164134
ENSDART00000024872 |
creb3l3l
|
cAMP responsive element binding protein 3-like 3 like |
| chr25_-_21894317 | 1.76 |
ENSDART00000089642
|
fbxo31
|
F-box protein 31 |
| chr12_+_13205955 | 1.75 |
ENSDART00000092906
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr25_+_26923193 | 1.64 |
ENSDART00000187364
|
grm8b
|
glutamate receptor, metabotropic 8b |
| chr10_+_10210455 | 1.61 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr24_-_37484123 | 1.56 |
ENSDART00000111623
|
cluap1
|
clusterin associated protein 1 |
| chr5_-_43935460 | 1.55 |
ENSDART00000166152
ENSDART00000188969 |
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr15_+_24737599 | 1.47 |
ENSDART00000078024
|
crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr24_-_38644937 | 1.47 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr16_+_10429770 | 1.46 |
ENSDART00000173132
|
vars
|
valyl-tRNA synthetase |
| chr15_+_45994123 | 1.44 |
ENSDART00000124704
|
lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr7_+_39664055 | 1.44 |
ENSDART00000146171
|
zgc:158564
|
zgc:158564 |
| chr16_+_4839078 | 1.43 |
ENSDART00000150111
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr3_+_13879446 | 1.31 |
ENSDART00000164767
|
farsa
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr20_+_28803977 | 1.28 |
ENSDART00000153351
ENSDART00000038149 |
fntb
|
farnesyltransferase, CAAX box, beta |
| chr14_-_47882706 | 1.22 |
ENSDART00000188772
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr4_+_16784553 | 1.17 |
ENSDART00000134054
|
si:dkey-13i19.8
|
si:dkey-13i19.8 |
| chr8_+_54202554 | 1.17 |
ENSDART00000020569
|
creld1b
|
cysteine-rich with EGF-like domains 1b |
| chr14_-_23801389 | 1.17 |
ENSDART00000054264
|
nr3c1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr3_+_40856095 | 1.16 |
ENSDART00000143207
|
mmd2a
|
monocyte to macrophage differentiation-associated 2a |
| chr20_-_14680897 | 1.11 |
ENSDART00000063857
ENSDART00000161314 |
scrn2
|
secernin 2 |
| chr20_-_32007209 | 1.09 |
ENSDART00000021575
|
adgb
|
androglobin |
| chr14_-_1454413 | 1.07 |
ENSDART00000185403
ENSDART00000191357 |
pmt
|
phosphoethanolamine methyltransferase |
| chr16_+_4838808 | 1.07 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr16_-_31351419 | 1.06 |
ENSDART00000178298
ENSDART00000018091 |
mroh1
|
maestro heat-like repeat family member 1 |
| chr7_+_38090515 | 1.00 |
ENSDART00000131387
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr8_+_7778770 | 0.98 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr2_-_32486080 | 0.97 |
ENSDART00000110821
|
ttc19
|
tetratricopeptide repeat domain 19 |
| chr23_-_43718067 | 0.97 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr22_-_20720427 | 0.97 |
ENSDART00000105532
|
oaz1a
|
ornithine decarboxylase antizyme 1a |
| chr13_+_22317216 | 0.95 |
ENSDART00000110794
|
usp54a
|
ubiquitin specific peptidase 54a |
| chr1_+_37312580 | 0.93 |
ENSDART00000171340
|
fam193a
|
family with sequence similarity 193, member A |
| chr18_+_27926839 | 0.92 |
ENSDART00000191835
|
hipk3b
|
homeodomain interacting protein kinase 3b |
| chr16_-_36093776 | 0.89 |
ENSDART00000159134
|
OSCP1
|
organic solute carrier partner 1 |
| chr14_+_7892383 | 0.86 |
ENSDART00000063837
|
ube2d2
|
ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) |
| chr3_-_15889508 | 0.84 |
ENSDART00000148363
|
cramp1
|
cramped chromatin regulator homolog 1 |
| chr15_-_33495048 | 0.83 |
ENSDART00000159882
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr14_-_1454045 | 0.82 |
ENSDART00000161460
|
pmt
|
phosphoethanolamine methyltransferase |
| chr8_+_46386601 | 0.81 |
ENSDART00000129661
ENSDART00000084081 |
ogg1
|
8-oxoguanine DNA glycosylase |
| chr18_-_2727764 | 0.80 |
ENSDART00000160841
|
ARHGEF17
|
si:ch211-248g20.5 |
| chr12_+_17603528 | 0.80 |
ENSDART00000111565
|
pms2
|
PMS1 homolog 2, mismatch repair system component |
| chr14_-_38809561 | 0.79 |
ENSDART00000159159
|
sil1
|
SIL1 nucleotide exchange factor |
| chr23_-_36441693 | 0.76 |
ENSDART00000024354
|
csad
|
cysteine sulfinic acid decarboxylase |
| chr14_+_7892021 | 0.73 |
ENSDART00000161307
|
ube2d2
|
ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) |
| chr22_+_38276024 | 0.73 |
ENSDART00000143792
|
rcor3
|
REST corepressor 3 |
| chr16_-_42770064 | 0.64 |
ENSDART00000112879
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr4_-_17642168 | 0.64 |
ENSDART00000007030
|
klhl42
|
kelch-like family, member 42 |
| chr5_-_43935119 | 0.60 |
ENSDART00000142271
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr16_+_41826584 | 0.46 |
ENSDART00000147523
|
si:dkey-199f5.6
|
si:dkey-199f5.6 |
| chr12_-_17602958 | 0.45 |
ENSDART00000134690
ENSDART00000028090 |
eif2ak1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr24_+_20536056 | 0.43 |
ENSDART00000082082
|
gars
|
glycyl-tRNA synthetase |
| chr3_+_19216567 | 0.39 |
ENSDART00000134433
|
il12rb2l
|
interleukin 12 receptor, beta 2a, like |
| chr13_-_9300299 | 0.37 |
ENSDART00000144142
|
HTRA2 (1 of many)
|
si:dkey-33c12.12 |
| chr15_-_23908605 | 0.37 |
ENSDART00000185578
|
usp32
|
ubiquitin specific peptidase 32 |
| chr19_-_8812891 | 0.34 |
ENSDART00000151134
ENSDART00000025385 ENSDART00000180291 |
cers2a
|
ceramide synthase 2a |
| chr4_+_12113105 | 0.33 |
ENSDART00000182399
|
tmem178b
|
transmembrane protein 178B |
| chr18_-_16922905 | 0.32 |
ENSDART00000187165
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr17_-_50220228 | 0.26 |
ENSDART00000058707
|
jdp2a
|
Jun dimerization protein 2a |
| chr9_+_747612 | 0.26 |
ENSDART00000181004
|
insig2
|
insulin induced gene 2 |
| chr22_-_24284447 | 0.26 |
ENSDART00000149894
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr15_-_24960730 | 0.19 |
ENSDART00000109990
ENSDART00000186706 |
abhd15a
|
abhydrolase domain containing 15a |
| chr12_+_17154655 | 0.13 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr15_-_5780593 | 0.13 |
ENSDART00000163555
ENSDART00000082388 |
chchd2
|
coiled-coil-helix-coiled-coil-helix domain containing 2 |
| chr3_+_1107102 | 0.11 |
ENSDART00000092690
|
srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr12_+_6065661 | 0.06 |
ENSDART00000142418
|
sgms1
|
sphingomyelin synthase 1 |
| chr25_-_6389713 | 0.02 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
Network of associatons between targets according to the STRING database.
First level regulatory network of atf4a+atf4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.6 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 2.9 | 11.6 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 2.6 | 7.7 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 2.5 | 12.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 2.4 | 7.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) regulation of protein activation cascade(GO:2000257) |
| 1.9 | 7.8 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 1.7 | 10.0 | GO:0090467 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 1.6 | 6.5 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 1.6 | 9.4 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 1.5 | 13.3 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 1.4 | 15.3 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 1.2 | 4.7 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 1.1 | 3.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 1.0 | 3.0 | GO:0019852 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 1.0 | 9.9 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.8 | 3.2 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.8 | 3.2 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.8 | 3.1 | GO:0019344 | homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.7 | 33.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.6 | 7.4 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.5 | 6.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.4 | 2.2 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 0.4 | 3.0 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.4 | 3.8 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.4 | 2.1 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.4 | 9.8 | GO:0051923 | sulfation(GO:0051923) |
| 0.3 | 8.0 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.3 | 5.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.3 | 1.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.3 | 1.9 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.3 | 1.2 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.3 | 6.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.3 | 3.5 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.2 | 2.8 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.2 | 2.0 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.2 | 1.9 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.2 | 1.0 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 6.0 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 3.3 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 8.9 | GO:0003146 | heart jogging(GO:0003146) |
| 0.1 | 1.8 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 14.4 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.1 | 2.2 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.1 | 3.2 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 3.8 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 3.1 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.1 | 0.4 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.3 | GO:0036315 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 1.9 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.1 | 2.7 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 1.0 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 1.6 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 1.6 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 0.8 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 4.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 1.0 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.1 | 0.6 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 1.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 3.0 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.1 | 0.3 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 1.0 | GO:0033753 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.1 | 1.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 1.6 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 1.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 3.9 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 4.3 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 1.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.9 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 2.7 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 2.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 15.7 | GO:0007420 | brain development(GO:0007420) |
| 0.0 | 1.8 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.8 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 4.8 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
| 0.0 | 0.1 | GO:0010885 | regulation of cholesterol storage(GO:0010885) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.5 | 3.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.3 | 1.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.3 | 20.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 5.1 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 9.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 7.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.2 | 3.0 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 6.3 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 15.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 7.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.8 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 1.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 3.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 4.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.0 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 3.5 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 2.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 3.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 3.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 17.3 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 18.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 14.2 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 2.9 | 11.6 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 1.5 | 7.7 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 1.4 | 10.0 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 1.4 | 15.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 1.3 | 6.5 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 1.0 | 4.8 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.9 | 3.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.9 | 9.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.8 | 9.9 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.8 | 3.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.8 | 3.8 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.6 | 15.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.5 | 2.0 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.4 | 1.8 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.4 | 2.6 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.4 | 3.0 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.4 | 2.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.4 | 3.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.3 | 1.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.3 | 3.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.3 | 6.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 1.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 1.6 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.2 | 1.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 5.1 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 0.8 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.2 | 2.7 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 8.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 2.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 2.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 11.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 9.8 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 4.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 33.1 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 4.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 3.3 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 1.9 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 4.7 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 2.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 2.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 4.7 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 3.5 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 18.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.2 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.0 | 1.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 3.2 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 5.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.8 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 4.0 | GO:0016887 | ATPase activity(GO:0016887) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 6.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 1.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 11.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 6.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 2.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 3.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 3.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 2.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 1.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 2.1 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 14.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 2.5 | 24.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 2.5 | 9.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 2.5 | 9.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 1.9 | 5.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.9 | 25.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.6 | 11.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.6 | 13.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.4 | 3.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 1.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.2 | 3.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 1.5 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 1.8 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 1.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.8 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 3.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |