Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
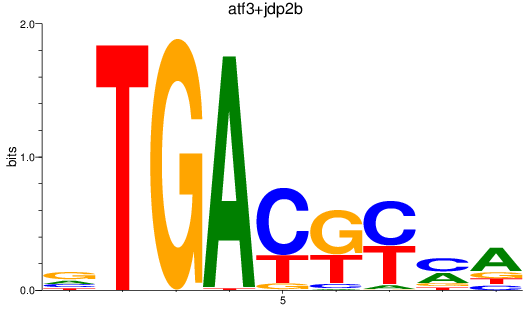
Results for atf3+jdp2b
Z-value: 1.93
Transcription factors associated with atf3+jdp2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
atf3
|
ENSDARG00000007823 | activating transcription factor 3 |
|
jdp2b
|
ENSDARG00000020133 | Jun dimerization protein 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| atf3 | dr11_v1_chr20_+_37794633_37794633 | 0.31 | 2.3e-03 | Click! |
| jdp2b | dr11_v1_chr20_+_46586678_46586692 | 0.26 | 1.2e-02 | Click! |
Activity profile of atf3+jdp2b motif
Sorted Z-values of atf3+jdp2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_112579 | 23.89 |
ENSDART00000034505
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr10_+_20256454 | 23.86 |
ENSDART00000111097
|
TCIM (1 of many)
|
transcriptional and immune response regulator |
| chr23_-_44723102 | 21.82 |
ENSDART00000129138
|
mogat3a
|
monoacylglycerol O-acyltransferase 3a |
| chr5_+_29831235 | 20.91 |
ENSDART00000109660
|
f11r.1
|
F11 receptor, tandem duplicate 1 |
| chr18_-_5598958 | 19.83 |
ENSDART00000161538
|
cyp1a
|
cytochrome P450, family 1, subfamily A |
| chr5_+_57658898 | 18.58 |
ENSDART00000074268
ENSDART00000124568 |
zgc:153929
|
zgc:153929 |
| chr24_-_26328721 | 18.52 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr14_+_21106444 | 16.97 |
ENSDART00000075744
ENSDART00000132363 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr5_-_29531948 | 16.48 |
ENSDART00000098360
|
arrdc1a
|
arrestin domain containing 1a |
| chr17_-_2039511 | 15.73 |
ENSDART00000160223
|
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr1_-_59240975 | 15.35 |
ENSDART00000166170
|
mvb12a
|
multivesicular body subunit 12A |
| chr14_-_11456724 | 15.31 |
ENSDART00000110424
|
si:ch211-153b23.4
|
si:ch211-153b23.4 |
| chr15_+_36054864 | 15.24 |
ENSDART00000156697
|
col4a3
|
collagen, type IV, alpha 3 |
| chr3_+_39540014 | 14.92 |
ENSDART00000074848
|
zgc:165423
|
zgc:165423 |
| chr17_+_30894431 | 14.41 |
ENSDART00000127996
|
degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr5_+_26213874 | 13.76 |
ENSDART00000193816
ENSDART00000098514 |
oclnb
|
occludin b |
| chr3_-_49110710 | 13.66 |
ENSDART00000160404
|
trim35-12
|
tripartite motif containing 35-12 |
| chr3_+_34670076 | 12.85 |
ENSDART00000133457
|
dlx4a
|
distal-less homeobox 4a |
| chr7_-_12968689 | 12.59 |
ENSDART00000173115
ENSDART00000013690 |
rplp2l
|
ribosomal protein, large P2, like |
| chr10_-_41352502 | 12.55 |
ENSDART00000052971
ENSDART00000128156 |
rab11fip1b
|
RAB11 family interacting protein 1 (class I) b |
| chr5_-_26181863 | 12.50 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr9_-_34269066 | 12.49 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr15_+_20239141 | 12.46 |
ENSDART00000101152
ENSDART00000152473 |
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr7_-_20241346 | 12.33 |
ENSDART00000173619
ENSDART00000127699 |
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr1_-_25679339 | 12.33 |
ENSDART00000161703
ENSDART00000054230 |
fgg
|
fibrinogen gamma chain |
| chr23_+_19790962 | 12.14 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr3_-_15734358 | 11.94 |
ENSDART00000137325
|
mvp
|
major vault protein |
| chr24_-_12938922 | 11.90 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr3_+_16922226 | 11.87 |
ENSDART00000017646
|
atp6v0a1a
|
ATPase H+ transporting V0 subunit a1a |
| chr2_-_22688651 | 11.77 |
ENSDART00000013863
|
agxtb
|
alanine-glyoxylate aminotransferase b |
| chr7_+_6652967 | 11.71 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr1_-_45157243 | 11.70 |
ENSDART00000131882
|
mucms1
|
mucin, multiple PTS and SEA group, member 1 |
| chr16_-_45178430 | 11.68 |
ENSDART00000165186
|
si:dkey-33i11.9
|
si:dkey-33i11.9 |
| chr5_-_69944084 | 11.67 |
ENSDART00000188557
ENSDART00000127782 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr22_+_1170294 | 11.61 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr18_+_2228737 | 11.43 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr24_-_40668208 | 11.42 |
ENSDART00000171543
|
smyhc1
|
slow myosin heavy chain 1 |
| chr20_-_35578435 | 11.35 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr14_+_11430796 | 11.22 |
ENSDART00000165275
|
si:ch211-153b23.3
|
si:ch211-153b23.3 |
| chr10_+_252425 | 11.21 |
ENSDART00000059478
|
lrrc32
|
leucine rich repeat containing 32 |
| chr19_-_977849 | 11.20 |
ENSDART00000172303
|
CABZ01088282.1
|
|
| chr22_-_10121880 | 10.93 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr25_+_37435720 | 10.79 |
ENSDART00000164390
|
chmp1a
|
charged multivesicular body protein 1A |
| chr24_-_33291784 | 10.71 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr10_-_322769 | 10.56 |
ENSDART00000165244
|
akt2l
|
v-akt murine thymoma viral oncogene homolog 2, like |
| chr25_+_31227747 | 10.55 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr3_-_19561058 | 10.54 |
ENSDART00000079323
|
zgc:163079
|
zgc:163079 |
| chr8_-_50259448 | 10.52 |
ENSDART00000146056
|
nkx3-1
|
NK3 homeobox 1 |
| chr5_+_45677781 | 10.45 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr3_+_42923275 | 10.33 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr7_+_33172066 | 10.11 |
ENSDART00000174013
|
si:ch211-194p6.12
|
si:ch211-194p6.12 |
| chr17_+_28102487 | 10.10 |
ENSDART00000131638
ENSDART00000076265 |
zgc:91908
|
zgc:91908 |
| chr15_+_36457888 | 10.08 |
ENSDART00000155100
|
si:dkey-262k9.2
|
si:dkey-262k9.2 |
| chr22_+_997838 | 9.95 |
ENSDART00000149743
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr3_+_26814030 | 9.86 |
ENSDART00000180128
|
socs1a
|
suppressor of cytokine signaling 1a |
| chr19_-_3781405 | 9.70 |
ENSDART00000170609
|
btr19
|
bloodthirsty-related gene family, member 19 |
| chr1_+_1896737 | 9.66 |
ENSDART00000152442
|
si:ch211-132g1.6
|
si:ch211-132g1.6 |
| chr24_-_40667800 | 9.59 |
ENSDART00000169315
|
smyhc1
|
slow myosin heavy chain 1 |
| chr1_-_59104145 | 9.55 |
ENSDART00000132495
ENSDART00000152457 |
MFAP4 (1 of many)
si:zfos-2330d3.7
|
si:zfos-2330d3.1 si:zfos-2330d3.7 |
| chr18_+_35842933 | 9.49 |
ENSDART00000151587
ENSDART00000131121 |
ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr16_+_33143503 | 9.45 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr19_+_14113886 | 9.24 |
ENSDART00000169343
|
kdf1b
|
keratinocyte differentiation factor 1b |
| chr24_+_35564668 | 9.23 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr16_+_42772678 | 9.23 |
ENSDART00000155575
|
si:ch211-135n15.2
|
si:ch211-135n15.2 |
| chr3_-_16784280 | 9.12 |
ENSDART00000137108
ENSDART00000137276 |
si:dkey-30j10.5
|
si:dkey-30j10.5 |
| chr22_-_10541372 | 9.07 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr4_-_13502549 | 9.03 |
ENSDART00000140366
|
si:ch211-266a5.12
|
si:ch211-266a5.12 |
| chr22_-_23666504 | 9.01 |
ENSDART00000158665
|
cfh
|
complement factor H |
| chr15_-_36055401 | 9.01 |
ENSDART00000154476
|
col4a4
|
collagen, type IV, alpha 4 |
| chr21_-_16219400 | 8.94 |
ENSDART00000124871
|
tnfrsfa
|
tumor necrosis factor receptor superfamily, member a |
| chr8_+_23725957 | 8.93 |
ENSDART00000104346
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr6_+_60055168 | 8.79 |
ENSDART00000008752
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr16_+_46725087 | 8.70 |
ENSDART00000008920
|
rab11al
|
RAB11a, member RAS oncogene family, like |
| chr22_-_22340688 | 8.67 |
ENSDART00000105597
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr3_-_61116258 | 8.64 |
ENSDART00000009194
ENSDART00000156978 |
aimp2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr10_-_15053507 | 8.58 |
ENSDART00000157446
ENSDART00000170441 |
CLDN23
|
si:ch211-95j8.5 |
| chr7_-_52417777 | 8.49 |
ENSDART00000110265
|
myzap
|
myocardial zonula adherens protein |
| chr7_+_38750871 | 8.43 |
ENSDART00000114238
ENSDART00000052325 ENSDART00000137001 |
f2
|
coagulation factor II (thrombin) |
| chr6_+_42475730 | 8.40 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr11_-_8167799 | 8.23 |
ENSDART00000133574
ENSDART00000024046 ENSDART00000146940 |
uox
|
urate oxidase |
| chr5_-_69934558 | 8.21 |
ENSDART00000124954
|
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr18_-_50799510 | 8.17 |
ENSDART00000174373
|
taldo1
|
transaldolase 1 |
| chr24_+_21621654 | 8.15 |
ENSDART00000002595
|
rpl21
|
ribosomal protein L21 |
| chr10_+_76864 | 8.11 |
ENSDART00000036375
|
vps26c
|
Down syndrome critical region 3 |
| chr4_-_74998614 | 8.09 |
ENSDART00000162529
|
zgc:172139
|
zgc:172139 |
| chr2_+_24762567 | 7.99 |
ENSDART00000078866
|
ifi30
|
interferon, gamma-inducible protein 30 |
| chr10_+_40792078 | 7.98 |
ENSDART00000140007
|
si:ch211-139n6.3
|
si:ch211-139n6.3 |
| chr13_+_646700 | 7.98 |
ENSDART00000006892
|
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr1_-_22678471 | 7.96 |
ENSDART00000128918
|
fgfbp1b
|
fibroblast growth factor binding protein 1b |
| chr2_-_55779927 | 7.96 |
ENSDART00000168579
|
CABZ01066725.1
|
|
| chr21_-_45382112 | 7.93 |
ENSDART00000151029
ENSDART00000151335 ENSDART00000151687 ENSDART00000075438 |
cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chr7_+_35268880 | 7.92 |
ENSDART00000182231
|
dpep2
|
dipeptidase 2 |
| chr2_-_32237916 | 7.91 |
ENSDART00000141418
|
fam49ba
|
family with sequence similarity 49, member Ba |
| chr1_-_23274038 | 7.87 |
ENSDART00000181658
|
rfc1
|
replication factor C (activator 1) 1 |
| chr23_+_28381260 | 7.87 |
ENSDART00000162722
|
zgc:153867
|
zgc:153867 |
| chr2_+_9560740 | 7.80 |
ENSDART00000003465
|
gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr21_+_45816030 | 7.66 |
ENSDART00000187056
|
pitx1
|
paired-like homeodomain 1 |
| chr22_+_26443235 | 7.65 |
ENSDART00000044085
|
zgc:92480
|
zgc:92480 |
| chr12_+_22588923 | 7.56 |
ENSDART00000184862
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr19_-_25005609 | 7.51 |
ENSDART00000151129
|
xkr8.2
|
XK, Kell blood group complex subunit-related family, member 8, tandem duplicate 2 |
| chr10_+_38643304 | 7.48 |
ENSDART00000067447
|
mmp30
|
matrix metallopeptidase 30 |
| chr24_+_39688072 | 7.43 |
ENSDART00000167900
|
zgc:153659
|
zgc:153659 |
| chr22_-_10156581 | 7.40 |
ENSDART00000168304
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr22_-_17781213 | 7.39 |
ENSDART00000137984
|
si:ch73-63e15.2
|
si:ch73-63e15.2 |
| chr24_-_39186185 | 7.39 |
ENSDART00000123019
ENSDART00000191114 |
nubp2
|
nucleotide binding protein 2 (MinD homolog, E. coli) |
| chr2_-_6482240 | 7.36 |
ENSDART00000132623
|
rgs13
|
regulator of G protein signaling 13 |
| chr12_-_49032899 | 7.36 |
ENSDART00000159356
|
GHITM
|
growth hormone inducible transmembrane protein |
| chr1_+_5402476 | 7.25 |
ENSDART00000040204
|
tuba8l2
|
tubulin, alpha 8 like 2 |
| chr13_-_17464362 | 7.21 |
ENSDART00000145499
|
lrmda
|
leucine rich melanocyte differentiation associated |
| chr24_+_37461457 | 7.15 |
ENSDART00000165775
|
nlrc3
|
NLR family, CARD domain containing 3 |
| chr24_+_10413484 | 7.08 |
ENSDART00000111014
|
myca
|
MYC proto-oncogene, bHLH transcription factor a |
| chr9_-_56231387 | 7.00 |
ENSDART00000149851
|
rpl31
|
ribosomal protein L31 |
| chr7_+_67451108 | 6.88 |
ENSDART00000163840
|
gcshb
|
glycine cleavage system protein H (aminomethyl carrier), b |
| chr23_+_34990693 | 6.87 |
ENSDART00000013449
|
CHST13
|
si:ch211-236h17.3 |
| chr22_-_15593824 | 6.86 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr14_+_39156 | 6.84 |
ENSDART00000082184
|
tmem107
|
transmembrane protein 107 |
| chr10_+_2600830 | 6.80 |
ENSDART00000101012
|
CU856539.2
|
|
| chr13_-_319996 | 6.80 |
ENSDART00000148675
|
ddx21
|
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr6_-_52675630 | 6.76 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr23_+_2421689 | 6.71 |
ENSDART00000180200
|
tcp1
|
t-complex 1 |
| chr4_-_77218637 | 6.71 |
ENSDART00000174325
|
psmb10
|
proteasome subunit beta 10 |
| chr3_-_33427803 | 6.68 |
ENSDART00000075495
|
rpl23
|
ribosomal protein L23 |
| chr8_-_36475328 | 6.68 |
ENSDART00000048448
|
si:busm1-266f07.2
|
si:busm1-266f07.2 |
| chr17_-_43666166 | 6.67 |
ENSDART00000077990
|
egr2a
|
early growth response 2a |
| chr3_-_7656059 | 6.65 |
ENSDART00000170917
|
junbb
|
JunB proto-oncogene, AP-1 transcription factor subunit b |
| chr3_-_1387292 | 6.57 |
ENSDART00000163535
|
ddx47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr18_+_6558338 | 6.53 |
ENSDART00000110892
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr25_+_6122823 | 6.53 |
ENSDART00000191824
ENSDART00000067514 |
rbpms2a
|
RNA binding protein with multiple splicing 2a |
| chr1_+_1689775 | 6.50 |
ENSDART00000048828
|
atp1a1a.4
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 4 |
| chr18_-_16953978 | 6.50 |
ENSDART00000100126
|
akip1
|
A kinase (PRKA) interacting protein 1 |
| chr10_-_1276046 | 6.48 |
ENSDART00000169779
|
pdlim5b
|
PDZ and LIM domain 5b |
| chr19_+_2590182 | 6.46 |
ENSDART00000162293
|
si:ch73-345f18.3
|
si:ch73-345f18.3 |
| chr1_-_44484 | 6.39 |
ENSDART00000171547
ENSDART00000164075 ENSDART00000168091 |
tmem39a
|
transmembrane protein 39A |
| chr8_-_25329967 | 6.39 |
ENSDART00000139682
|
eps8l3b
|
EPS8-like 3b |
| chr1_-_23274393 | 6.33 |
ENSDART00000147800
ENSDART00000130277 ENSDART00000054340 ENSDART00000054338 |
rpl9
|
ribosomal protein L9 |
| chr2_+_40181633 | 6.32 |
ENSDART00000185494
|
FP236331.1
|
|
| chr12_-_9132682 | 6.31 |
ENSDART00000066471
|
adam8b
|
ADAM metallopeptidase domain 8b |
| chr25_-_23526058 | 6.31 |
ENSDART00000191331
ENSDART00000062930 |
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr3_-_19367081 | 6.31 |
ENSDART00000191369
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr2_-_37280617 | 6.26 |
ENSDART00000190458
|
nadkb
|
NAD kinase b |
| chr13_+_233482 | 6.26 |
ENSDART00000102511
|
cfap36
|
cilia and flagella associated protein 36 |
| chr8_-_38317914 | 6.22 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr25_-_35182347 | 6.21 |
ENSDART00000115210
|
ano9a
|
anoctamin 9a |
| chr20_-_54245256 | 6.19 |
ENSDART00000170482
|
prpf39
|
PRP39 pre-mRNA processing factor 39 homolog (yeast) |
| chr14_-_12822 | 6.19 |
ENSDART00000180650
ENSDART00000188819 |
msx1a
|
muscle segment homeobox 1a |
| chr15_-_37104165 | 6.18 |
ENSDART00000165867
|
zmp:0000001114
|
zmp:0000001114 |
| chr7_+_10592152 | 6.18 |
ENSDART00000182624
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr13_-_41908583 | 6.18 |
ENSDART00000136515
|
ipmka
|
inositol polyphosphate multikinase a |
| chr15_-_41689684 | 6.15 |
ENSDART00000143447
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr2_+_52232630 | 6.15 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr5_+_27898226 | 6.15 |
ENSDART00000098604
ENSDART00000180251 |
adam28
|
ADAM metallopeptidase domain 28 |
| chr20_+_36812368 | 6.09 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr16_+_21790870 | 6.07 |
ENSDART00000155039
|
trim108
|
tripartite motif containing 108 |
| chr4_-_73190246 | 6.04 |
ENSDART00000170842
|
LO018260.1
|
|
| chr23_-_10722664 | 6.03 |
ENSDART00000146526
ENSDART00000129022 ENSDART00000104985 |
foxp1a
|
forkhead box P1a |
| chr7_-_19168375 | 6.02 |
ENSDART00000112447
|
il13ra1
|
interleukin 13 receptor, alpha 1 |
| chr17_-_32370047 | 6.01 |
ENSDART00000145487
|
klf11b
|
Kruppel-like factor 11b |
| chr24_+_37406535 | 5.95 |
ENSDART00000138264
|
si:ch211-183d21.1
|
si:ch211-183d21.1 |
| chr25_+_753364 | 5.94 |
ENSDART00000183804
|
TWF1 (1 of many)
|
twinfilin actin binding protein 1 |
| chr19_-_25081711 | 5.92 |
ENSDART00000058513
|
xkr8.3
|
XK, Kell blood group complex subunit-related family, member 8, tandem duplicate 3 |
| chr18_+_48423973 | 5.92 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr10_+_4924388 | 5.91 |
ENSDART00000108595
|
slc46a2
|
solute carrier family 46 member 2 |
| chr25_+_3099073 | 5.89 |
ENSDART00000022506
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr15_-_8309207 | 5.86 |
ENSDART00000143880
ENSDART00000061351 |
tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr15_-_41689981 | 5.83 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr5_-_3991655 | 5.83 |
ENSDART00000159368
|
myo19
|
myosin XIX |
| chr16_+_20947439 | 5.83 |
ENSDART00000137344
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr7_+_41296795 | 5.82 |
ENSDART00000173562
|
si:dkey-86l18.10
|
si:dkey-86l18.10 |
| chr10_-_14488472 | 5.81 |
ENSDART00000101298
ENSDART00000138161 |
galt
|
galactose-1-phosphate uridylyltransferase |
| chr6_+_13201358 | 5.81 |
ENSDART00000190290
|
CT009620.1
|
|
| chr17_+_15388479 | 5.81 |
ENSDART00000052439
|
si:ch211-266g18.6
|
si:ch211-266g18.6 |
| chr19_-_2231146 | 5.79 |
ENSDART00000181909
ENSDART00000043595 |
twist1a
|
twist family bHLH transcription factor 1a |
| chr24_-_27436319 | 5.76 |
ENSDART00000171489
|
si:dkey-25o1.7
|
si:dkey-25o1.7 |
| chr24_+_7828613 | 5.75 |
ENSDART00000135161
|
zgc:101569
|
zgc:101569 |
| chr7_-_24364536 | 5.74 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr13_-_11971148 | 5.74 |
ENSDART00000066230
ENSDART00000185614 |
ARL3 (1 of many)
|
zgc:110197 |
| chr19_-_12404590 | 5.71 |
ENSDART00000103703
|
ftr56
|
finTRIM family, member 56 |
| chr2_-_37280028 | 5.62 |
ENSDART00000139459
|
nadkb
|
NAD kinase b |
| chr1_+_11659861 | 5.60 |
ENSDART00000054787
|
BX569798.1
|
|
| chr8_+_52415603 | 5.59 |
ENSDART00000021604
ENSDART00000191424 |
gins4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr11_+_16216909 | 5.58 |
ENSDART00000081035
ENSDART00000147190 |
slc25a26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr25_-_6049339 | 5.57 |
ENSDART00000075184
|
snx1a
|
sorting nexin 1a |
| chr19_+_10603405 | 5.55 |
ENSDART00000151135
|
si:dkey-211g8.8
|
si:dkey-211g8.8 |
| chr2_+_36121373 | 5.55 |
ENSDART00000187002
|
CT867973.2
|
|
| chr4_+_8532580 | 5.53 |
ENSDART00000162065
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr5_-_28016805 | 5.52 |
ENSDART00000078642
|
vps37b
|
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
| chr2_-_30721502 | 5.51 |
ENSDART00000132389
|
si:dkey-94e7.1
|
si:dkey-94e7.1 |
| chr23_-_33738945 | 5.49 |
ENSDART00000136386
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr20_+_46925581 | 5.49 |
ENSDART00000192531
|
si:ch73-21k16.5
|
si:ch73-21k16.5 |
| chr5_+_64856666 | 5.49 |
ENSDART00000050863
|
zgc:101858
|
zgc:101858 |
| chr4_+_77948970 | 5.48 |
ENSDART00000149636
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr8_-_65189 | 5.48 |
ENSDART00000168412
|
hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr22_-_607812 | 5.46 |
ENSDART00000145983
|
cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr5_+_72145468 | 5.45 |
ENSDART00000148626
|
abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr8_-_43775805 | 5.44 |
ENSDART00000090534
ENSDART00000144239 |
ulk1a
|
unc-51 like autophagy activating kinase 1a |
| chr23_+_44049509 | 5.43 |
ENSDART00000102003
|
txk
|
TXK tyrosine kinase |
| chr12_+_45238292 | 5.41 |
ENSDART00000057983
|
mrpl38
|
mitochondrial ribosomal protein L38 |
| chr13_-_36582341 | 5.41 |
ENSDART00000137335
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr19_+_43341424 | 5.40 |
ENSDART00000134815
|
sesn2
|
sestrin 2 |
| chr21_+_20383837 | 5.40 |
ENSDART00000026430
|
hspb11
|
heat shock protein, alpha-crystallin-related, b11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of atf3+jdp2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 20.7 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 5.1 | 15.4 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 3.0 | 21.0 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 3.0 | 30.0 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 2.7 | 8.2 | GO:0046415 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 2.7 | 16.0 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 2.5 | 12.7 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 2.4 | 2.4 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 2.2 | 8.9 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 2.2 | 2.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 2.1 | 6.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 2.0 | 2.0 | GO:1903430 | negative regulation of cell maturation(GO:1903430) |
| 1.8 | 10.9 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 1.7 | 8.7 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 1.7 | 6.9 | GO:0006544 | glycine metabolic process(GO:0006544) glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 1.7 | 11.9 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 1.7 | 6.8 | GO:0048103 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 1.6 | 24.7 | GO:0090559 | regulation of membrane permeability(GO:0090559) |
| 1.6 | 9.9 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 1.6 | 8.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 1.6 | 4.9 | GO:0036363 | transforming growth factor beta activation(GO:0036363) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) |
| 1.5 | 4.6 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 1.5 | 4.5 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 1.5 | 6.0 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 1.5 | 7.5 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 1.5 | 7.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 1.4 | 10.1 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 1.4 | 7.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 1.4 | 2.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 1.3 | 4.0 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 1.3 | 5.4 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 1.3 | 6.5 | GO:0051148 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 1.3 | 3.8 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 1.3 | 17.9 | GO:0034311 | diol metabolic process(GO:0034311) |
| 1.2 | 3.7 | GO:0014809 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.2 | 32.0 | GO:0030168 | platelet activation(GO:0030168) |
| 1.2 | 3.7 | GO:2000374 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 1.2 | 5.9 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 1.2 | 4.7 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 1.2 | 4.7 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 1.2 | 5.8 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 1.1 | 6.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 1.1 | 5.5 | GO:1904105 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 1.1 | 9.9 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 1.1 | 7.6 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 1.1 | 3.2 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 1.0 | 6.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 1.0 | 6.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 1.0 | 6.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 1.0 | 3.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 1.0 | 4.0 | GO:0032534 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 1.0 | 12.6 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 1.0 | 3.9 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 1.0 | 2.9 | GO:0097435 | fibril organization(GO:0097435) |
| 1.0 | 2.9 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) |
| 0.9 | 4.7 | GO:0051103 | DNA ligation(GO:0006266) immunoglobulin V(D)J recombination(GO:0033152) DNA ligation involved in DNA repair(GO:0051103) |
| 0.9 | 5.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.9 | 4.6 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.9 | 13.8 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.9 | 11.9 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.9 | 2.7 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.9 | 5.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.9 | 7.2 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.9 | 2.6 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.9 | 3.4 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.8 | 3.4 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.8 | 2.5 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.8 | 3.4 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.8 | 10.8 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.8 | 30.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.8 | 3.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.8 | 5.4 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.8 | 5.4 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.8 | 3.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.8 | 11.3 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.7 | 2.2 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.7 | 8.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.7 | 2.2 | GO:0032239 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.7 | 3.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.7 | 5.1 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.7 | 3.6 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.7 | 2.1 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.7 | 4.9 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.7 | 3.5 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.7 | 2.1 | GO:0090266 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.7 | 4.9 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.7 | 9.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.7 | 7.6 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.7 | 21.8 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.7 | 4.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.7 | 2.0 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.7 | 2.6 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.7 | 11.1 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.7 | 2.0 | GO:0035194 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.7 | 6.5 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.6 | 1.9 | GO:2000793 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.6 | 1.9 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.6 | 6.8 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.6 | 6.2 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.6 | 2.5 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.6 | 2.5 | GO:0051645 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.6 | 6.7 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.6 | 3.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.6 | 6.0 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.6 | 1.8 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.6 | 9.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.6 | 10.7 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.6 | 1.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.6 | 16.1 | GO:0007568 | aging(GO:0007568) |
| 0.6 | 4.6 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.6 | 4.5 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.6 | 3.4 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.6 | 1.7 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.5 | 5.5 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.5 | 1.6 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.5 | 1.6 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.5 | 3.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.5 | 4.2 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.5 | 2.6 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.5 | 2.0 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.5 | 2.5 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.5 | 10.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.5 | 4.0 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.5 | 2.0 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.5 | 2.0 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.5 | 3.9 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.5 | 11.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.5 | 2.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.5 | 5.4 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.5 | 1.5 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.5 | 3.3 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.5 | 3.2 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.4 | 4.0 | GO:0042987 | Notch receptor processing(GO:0007220) beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.4 | 14.0 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.4 | 23.1 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.4 | 7.4 | GO:0051340 | regulation of ligase activity(GO:0051340) |
| 0.4 | 2.1 | GO:0090200 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.4 | 1.3 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.4 | 3.4 | GO:1902808 | positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.4 | 1.2 | GO:0072540 | response to protozoan(GO:0001562) T-helper cell lineage commitment(GO:0002295) T cell lineage commitment(GO:0002360) alpha-beta T cell lineage commitment(GO:0002363) defense response to protozoan(GO:0042832) CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.4 | 2.5 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.4 | 1.6 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.4 | 9.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.4 | 10.0 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.4 | 3.0 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.4 | 7.2 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.4 | 2.6 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.4 | 5.6 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.4 | 1.9 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.4 | 3.7 | GO:0000479 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.4 | 1.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.4 | 0.4 | GO:0044819 | mitotic G1/S transition checkpoint(GO:0044819) |
| 0.4 | 1.8 | GO:0071333 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.4 | 0.7 | GO:0016246 | RNA interference(GO:0016246) |
| 0.3 | 3.8 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.3 | 3.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.3 | 2.4 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.3 | 6.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.3 | 6.0 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.3 | 5.6 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.3 | 2.0 | GO:0014004 | microglia differentiation(GO:0014004) |
| 0.3 | 1.9 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.3 | 1.6 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.3 | 1.3 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.3 | 0.9 | GO:0036315 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.3 | 2.8 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.3 | 1.6 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.3 | 7.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.3 | 6.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.3 | 6.2 | GO:0003181 | atrioventricular valve morphogenesis(GO:0003181) |
| 0.3 | 7.7 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.3 | 5.4 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.3 | 3.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 | 0.9 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.3 | 7.2 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.3 | 1.4 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.3 | 2.6 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.3 | 3.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.3 | 1.9 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.3 | 6.9 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.3 | 2.7 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.3 | 7.9 | GO:1903039 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.3 | 1.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 2.7 | GO:0010889 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.3 | 2.4 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.3 | 2.6 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 1.6 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.3 | 6.0 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.3 | 1.5 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.3 | 1.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.3 | 4.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.3 | 2.5 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 | 2.0 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 2.4 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.2 | 7.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.2 | 7.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.2 | 3.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 5.3 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.2 | 1.4 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 5.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.2 | 7.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 4.0 | GO:0032392 | DNA geometric change(GO:0032392) DNA duplex unwinding(GO:0032508) |
| 0.2 | 11.9 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.2 | 0.2 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.2 | 2.0 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.2 | 1.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 1.7 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.2 | 2.1 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.2 | 0.9 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.2 | 6.8 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.2 | 2.7 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.2 | 3.1 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine-containing compound transmembrane transport(GO:0072531) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.2 | 1.0 | GO:0060339 | regulation of type I interferon-mediated signaling pathway(GO:0060338) negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.2 | 0.6 | GO:0070589 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.2 | 2.0 | GO:0007584 | response to nutrient(GO:0007584) |
| 0.2 | 1.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.2 | 0.8 | GO:0071632 | optomotor response(GO:0071632) |
| 0.2 | 3.8 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.2 | 6.5 | GO:0050727 | regulation of inflammatory response(GO:0050727) |
| 0.2 | 2.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 3.5 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.2 | 20.2 | GO:0071407 | cellular response to organic cyclic compound(GO:0071407) |
| 0.2 | 2.7 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.2 | 16.3 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.2 | 1.5 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.2 | 5.8 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.2 | 1.8 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.2 | 4.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 2.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.2 | 3.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 3.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.2 | 2.7 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.2 | 2.5 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.2 | 1.7 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.2 | 14.8 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.2 | 4.0 | GO:0043588 | skin development(GO:0043588) |
| 0.2 | 0.8 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.2 | 2.8 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) regulation of polysaccharide biosynthetic process(GO:0032885) |
| 0.2 | 1.5 | GO:0014028 | notochord formation(GO:0014028) |
| 0.2 | 9.4 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.2 | 3.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 4.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.2 | 1.7 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 5.5 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.2 | 2.5 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.2 | 12.4 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.2 | 8.0 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 1.3 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 4.2 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.1 | 1.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.7 | GO:0046102 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.1 | 5.8 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.1 | 4.3 | GO:2001236 | regulation of extrinsic apoptotic signaling pathway(GO:2001236) |
| 0.1 | 0.6 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 6.6 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.1 | 0.4 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.1 | 3.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 4.4 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.1 | 10.1 | GO:0042113 | B cell activation(GO:0042113) |
| 0.1 | 2.8 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 4.4 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 2.8 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 | 2.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 8.2 | GO:0009306 | protein secretion(GO:0009306) |
| 0.1 | 0.2 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 3.1 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 3.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 3.8 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 4.3 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.1 | 0.9 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.1 | 1.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 8.9 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.1 | 6.7 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.1 | 3.0 | GO:0035272 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.1 | 6.0 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 1.6 | GO:0009409 | response to cold(GO:0009409) positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 4.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 0.7 | GO:1905168 | positive regulation of DNA recombination(GO:0045911) positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.1 | 1.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 2.1 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 0.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.6 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 2.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 17.1 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 1.2 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 0.6 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 1.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 3.6 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.1 | 6.0 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 1.8 | GO:0072663 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 10.2 | GO:0061053 | somite development(GO:0061053) |
| 0.1 | 3.2 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.1 | 1.8 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 1.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 7.0 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.1 | 6.3 | GO:0043068 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.1 | 3.8 | GO:1902652 | cholesterol metabolic process(GO:0008203) secondary alcohol metabolic process(GO:1902652) |
| 0.1 | 5.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 5.3 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.1 | 1.7 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.1 | 1.0 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 5.1 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 0.9 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.1 | 0.3 | GO:0060544 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 3.9 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.1 | 3.2 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.1 | 0.8 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 0.9 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 2.8 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 3.5 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.1 | 2.2 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.1 | 0.5 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 1.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 1.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 22.3 | GO:0006412 | translation(GO:0006412) |
| 0.1 | 3.7 | GO:0045765 | regulation of angiogenesis(GO:0045765) |
| 0.1 | 2.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 3.2 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 2.6 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 3.7 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.1 | 1.5 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 3.4 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.1 | 0.5 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 2.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 10.8 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.1 | 1.0 | GO:0035924 | cellular response to vascular endothelial growth factor stimulus(GO:0035924) |
| 0.1 | 1.1 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.1 | 30.8 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.1 | 0.1 | GO:0006458 | 'de novo' protein folding(GO:0006458) 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.5 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 1.1 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 1.4 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.3 | GO:0001776 | leukocyte homeostasis(GO:0001776) |
| 0.0 | 2.1 | GO:0048278 | membrane docking(GO:0022406) vesicle docking(GO:0048278) |
| 0.0 | 0.6 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 4.9 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.1 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.0 | 15.0 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 2.9 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 2.4 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.2 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 25.2 | GO:0006508 | proteolysis(GO:0006508) |
| 0.0 | 5.6 | GO:0061061 | muscle structure development(GO:0061061) |
| 0.0 | 0.3 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 3.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 2.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.4 | GO:0031333 | negative regulation of protein complex assembly(GO:0031333) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 1.8 | GO:0045165 | cell fate commitment(GO:0045165) |
| 0.0 | 0.1 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.2 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 1.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 4.5 | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter(GO:0045944) |
| 0.0 | 1.0 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.5 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 2.3 | 6.9 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 2.1 | 6.3 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 1.8 | 5.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 1.6 | 4.7 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 1.5 | 4.5 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 1.5 | 5.9 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 1.3 | 5.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 1.3 | 10.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 1.1 | 5.6 | GO:0000811 | GINS complex(GO:0000811) |
| 1.0 | 16.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.9 | 7.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.9 | 6.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.9 | 1.7 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.8 | 23.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.8 | 10.3 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) |
| 0.8 | 3.8 | GO:0055087 | Ski complex(GO:0055087) |
| 0.8 | 12.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.8 | 6.8 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.7 | 8.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.7 | 2.9 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.7 | 2.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.7 | 3.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.7 | 4.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.7 | 4.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.7 | 5.4 | GO:0000796 | condensin complex(GO:0000796) |
| 0.7 | 2.0 | GO:1990077 | primosome complex(GO:1990077) |
| 0.6 | 6.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.6 | 2.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.6 | 4.3 | GO:0016234 | inclusion body(GO:0016234) |
| 0.6 | 1.8 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.6 | 4.8 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.6 | 10.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.6 | 1.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.6 | 3.9 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.5 | 3.8 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.5 | 3.8 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.5 | 3.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.5 | 4.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.5 | 3.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.5 | 5.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.5 | 7.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.5 | 10.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.5 | 17.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.5 | 2.8 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.5 | 6.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.4 | 5.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.4 | 25.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.4 | 4.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.4 | 16.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.4 | 23.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.3 | 32.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 3.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.3 | 6.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.3 | 0.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 14.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.3 | 1.5 | GO:0043198 | ciliary rootlet(GO:0035253) dendritic shaft(GO:0043198) |
| 0.3 | 37.7 | GO:0005840 | ribosome(GO:0005840) |
| 0.3 | 2.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.3 | 3.5 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.3 | 1.7 | GO:0016589 | NURF complex(GO:0016589) |
| 0.3 | 4.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.3 | 3.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.3 | 0.9 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.3 | 2.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.3 | 2.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.3 | 3.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 7.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.3 | 3.9 | GO:0070187 | telosome(GO:0070187) |
| 0.3 | 11.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.3 | 28.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.3 | 30.9 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.2 | 10.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 1.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 1.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 1.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 12.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 18.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 1.0 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 23.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 1.3 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.2 | 3.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.2 | 2.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.2 | 2.5 | GO:0071010 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.2 | 2.2 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.2 | 2.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 2.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 19.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 0.6 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.7 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 6.0 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 4.5 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 2.9 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 2.7 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 2.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 10.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 1.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 2.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.6 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 2.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 5.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.6 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 3.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 3.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.9 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 2.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 65.0 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 7.8 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 5.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 25.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.1 | 0.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.8 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 3.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 4.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.4 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 0.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.3 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 1.0 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 21.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 1.2 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 0.2 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 1.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 7.2 | GO:0000786 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.1 | 2.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.3 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 0.8 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 2.0 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 0.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 16.3 | GO:0005764 | lysosome(GO:0005764) |
| 0.1 | 2.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 0.2 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 17.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 1.7 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 4.0 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.1 | 1.5 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 3.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 43.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 13.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 5.3 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0032806 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) carboxy-terminal domain protein kinase complex(GO:0032806) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.4 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 16.3 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) translation preinitiation complex(GO:0070993) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 1.6 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 3.3 | 19.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 3.3 | 16.5 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 2.2 | 10.9 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 2.2 | 21.8 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 2.1 | 17.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 2.1 | 6.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 1.7 | 6.8 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 1.6 | 7.9 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 1.5 | 4.6 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 1.5 | 4.5 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 1.5 | 7.5 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 1.5 | 8.9 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 1.5 | 11.7 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 1.5 | 4.4 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 1.4 | 10.1 | GO:0070728 | leucine binding(GO:0070728) |
| 1.4 | 4.2 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 1.4 | 8.2 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 1.4 | 4.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 1.3 | 11.7 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 1.2 | 6.2 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 1.2 | 3.7 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 1.2 | 4.8 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 1.2 | 4.7 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 1.1 | 4.5 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 1.1 | 8.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 1.0 | 3.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 1.0 | 8.2 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 1.0 | 5.0 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 1.0 | 3.9 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 1.0 | 22.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 1.0 | 7.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 1.0 | 4.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 1.0 | 4.8 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.9 | 5.6 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.9 | 3.7 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.9 | 5.5 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.9 | 4.3 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.9 | 2.6 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.8 | 2.4 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.7 | 3.6 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.7 | 2.2 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.7 | 4.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.7 | 7.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.7 | 2.1 | GO:0032138 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.7 | 3.5 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.7 | 4.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.7 | 2.0 | GO:0015562 | inositol hexakisphosphate binding(GO:0000822) efflux transmembrane transporter activity(GO:0015562) |
| 0.6 | 1.9 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.6 | 3.9 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.6 | 12.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.6 | 1.8 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.6 | 27.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.6 | 4.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.6 | 2.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.6 | 4.5 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.6 | 2.8 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.5 | 1.6 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.5 | 4.8 | GO:0097001 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.5 | 5.8 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.5 | 4.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.5 | 21.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.5 | 1.5 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.5 | 5.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.5 | 2.5 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.5 | 11.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.5 | 3.9 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.5 | 3.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.4 | 4.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.4 | 8.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.4 | 2.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.4 | 2.6 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.4 | 5.1 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.4 | 24.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.4 | 2.5 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.4 | 1.6 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.4 | 6.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.4 | 10.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.4 | 2.0 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.4 | 2.0 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.4 | 1.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.4 | 1.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.4 | 2.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.4 | 6.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 28.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.3 | 3.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.3 | 3.8 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.3 | 1.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 5.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.3 | 5.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 1.0 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.3 | 3.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.3 | 61.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.3 | 1.3 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.3 | 1.6 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.3 | 3.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.3 | 10.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.3 | 69.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.3 | 3.0 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.3 | 5.0 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.3 | 2.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.3 | 3.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.3 | 5.6 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.3 | 2.2 | GO:0001032 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.3 | 1.9 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.3 | 9.9 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.3 | 3.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.3 | 7.9 | GO:0099604 | ligand-gated calcium channel activity(GO:0099604) |
| 0.3 | 0.8 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.3 | 34.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.3 | 0.8 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.3 | 28.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 1.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.3 | 0.8 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.2 | 2.9 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 3.4 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.2 | 2.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.2 | 3.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.2 | 6.9 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.2 | 6.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 2.6 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.2 | 18.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 1.6 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 9.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 1.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 1.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.2 | 2.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 0.9 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.2 | 20.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 1.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 2.7 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.2 | 1.2 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.2 | 0.8 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 2.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 2.4 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.2 | 2.8 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 3.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 12.2 | GO:0003823 | antigen binding(GO:0003823) |
| 0.2 | 25.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 3.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 2.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 3.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 3.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 1.5 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.2 | 2.4 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.2 | 0.8 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.2 | 2.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 2.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.2 | 1.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 3.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 12.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 5.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.5 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 1.6 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 1.1 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.1 | 3.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.6 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 7.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 3.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 2.9 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 3.8 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 3.6 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 5.8 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.6 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 3.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.6 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.1 | 6.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 7.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 3.9 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 2.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.7 | GO:0043566 | structure-specific DNA binding(GO:0043566) |
| 0.1 | 1.9 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.2 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.1 | 1.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 22.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 2.7 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 0.6 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 4.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 2.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 1.3 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.1 | 1.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 6.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 0.7 | GO:0043394 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.1 | 5.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.6 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.1 | 1.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 4.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 13.2 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.1 | 2.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 1.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 2.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 2.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 22.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.6 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 3.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.7 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.1 | 2.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.4 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.1 | 23.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 2.9 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.1 | 0.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.8 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 1.1 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.1 | 0.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 6.0 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 1.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 3.0 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 1.0 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 1.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.9 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 2.5 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 2.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 4.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.5 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 2.9 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 2.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 1.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 3.2 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 1.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 1.3 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 1.0 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase activity(GO:0003916) DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 6.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.5 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 42.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 1.0 | 43.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.8 | 6.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.5 | 15.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.5 | 5.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 6.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.4 | 11.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.4 | 10.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.4 | 20.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.4 | 2.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.3 | 2.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.3 | 5.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.3 | 5.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.3 | 8.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.3 | 8.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.3 | 2.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 6.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.3 | 5.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.3 | 3.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.3 | 2.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 2.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.3 | 3.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.3 | 2.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 9.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.3 | 6.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 9.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 3.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 7.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 8.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 3.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 4.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.2 | 2.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 3.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 4.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.2 | 3.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.2 | 1.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 2.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 0.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 4.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 2.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 2.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 2.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 11.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 2.3 | 9.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 2.2 | 24.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 2.1 | 10.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.9 | 20.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.2 | 26.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 1.0 | 5.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.8 | 4.7 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.8 | 5.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.7 | 5.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.7 | 13.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.7 | 24.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.6 | 7.0 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.6 | 17.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.6 | 15.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.6 | 6.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.5 | 11.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.5 | 6.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.5 | 8.6 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.5 | 7.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.5 | 10.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.5 | 3.6 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.4 | 46.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.4 | 5.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.4 | 5.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.4 | 2.0 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.4 | 1.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.4 | 4.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.4 | 6.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.4 | 10.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.4 | 10.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.3 | 5.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.3 | 5.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.3 | 6.7 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.3 | 3.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.3 | 2.5 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.3 | 2.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 2.0 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.3 | 3.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 3.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.3 | 7.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 3.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.2 | 10.4 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.2 | 6.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 3.0 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 1.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 0.9 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.2 | 11.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 5.3 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 4.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 2.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.5 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 5.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.4 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 2.9 | REACTOME GLOBAL GENOMIC NER GG NER | Genes involved in Global Genomic NER (GG-NER) |
| 0.1 | 2.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 6.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 3.6 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.1 | 1.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 2.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.6 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 2.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 4.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 4.3 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.1 | 1.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.0 | REACTOME DNA REPLICATION | Genes involved in DNA Replication |
| 0.1 | 0.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 5.1 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 1.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.5 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |