Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
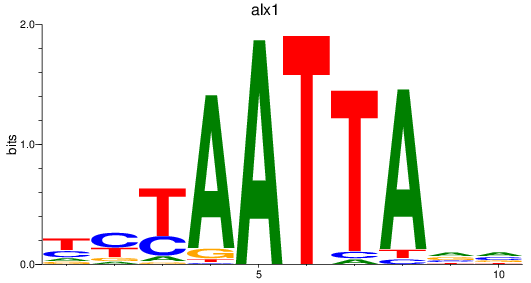
Results for alx1
Z-value: 1.19
Transcription factors associated with alx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
alx1
|
ENSDARG00000062824 | ALX homeobox 1 |
|
alx1
|
ENSDARG00000110530 | ALX homeobox 1 |
|
alx1
|
ENSDARG00000115230 | ALX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| alx1 | dr11_v1_chr18_+_16246806_16246806 | -0.13 | 2.0e-01 | Click! |
Activity profile of alx1 motif
Sorted Z-values of alx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_31276842 | 48.58 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr6_+_55032439 | 17.03 |
ENSDART00000164232
ENSDART00000158845 ENSDART00000157584 ENSDART00000026359 ENSDART00000122794 ENSDART00000183742 |
mybphb
|
myosin binding protein Hb |
| chr25_+_31277415 | 13.01 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr13_+_2338761 | 12.70 |
ENSDART00000102791
ENSDART00000129911 |
klhl31
|
kelch-like family member 31 |
| chr5_+_51594209 | 12.36 |
ENSDART00000164668
ENSDART00000058403 ENSDART00000055857 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr25_+_31267268 | 12.21 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr25_+_3306620 | 11.26 |
ENSDART00000182085
ENSDART00000034704 |
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr21_-_22737228 | 9.92 |
ENSDART00000151366
|
fbxo40.2
|
F-box protein 40, tandem duplicate 2 |
| chr16_-_17197546 | 9.90 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr21_-_23110841 | 9.39 |
ENSDART00000147896
ENSDART00000003076 ENSDART00000184925 ENSDART00000190386 |
usp28
|
ubiquitin specific peptidase 28 |
| chr9_-_22821901 | 8.69 |
ENSDART00000101711
|
neb
|
nebulin |
| chr14_+_22113331 | 8.67 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr10_-_21362320 | 8.40 |
ENSDART00000189789
|
avd
|
avidin |
| chr23_-_27571667 | 8.10 |
ENSDART00000008174
|
pfkma
|
phosphofructokinase, muscle a |
| chr9_-_22834860 | 7.81 |
ENSDART00000146486
|
neb
|
nebulin |
| chr6_-_54826061 | 7.63 |
ENSDART00000149982
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr10_-_21362071 | 7.39 |
ENSDART00000125167
|
avd
|
avidin |
| chr19_+_15441022 | 7.38 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr2_-_30324610 | 7.09 |
ENSDART00000185422
|
jph1b
|
junctophilin 1b |
| chr6_+_23887314 | 6.75 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr19_+_46158078 | 6.59 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr2_-_30324297 | 6.36 |
ENSDART00000099078
|
jph1b
|
junctophilin 1b |
| chr7_-_51773166 | 6.32 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr9_-_22822084 | 6.28 |
ENSDART00000142020
|
neb
|
nebulin |
| chr2_-_15324837 | 6.21 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr8_+_45334255 | 6.08 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr7_+_20471315 | 5.95 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr9_+_8396755 | 5.93 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr19_+_40983221 | 5.80 |
ENSDART00000144544
|
col1a2
|
collagen, type I, alpha 2 |
| chr6_-_46861676 | 5.78 |
ENSDART00000188712
ENSDART00000190148 |
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr24_+_17270129 | 5.60 |
ENSDART00000186729
|
spag6
|
sperm associated antigen 6 |
| chr9_-_49493305 | 5.60 |
ENSDART00000148707
ENSDART00000148561 |
xirp2b
|
xin actin binding repeat containing 2b |
| chr19_+_15440841 | 5.53 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr5_-_41494831 | 5.30 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr18_+_3037998 | 5.30 |
ENSDART00000185844
ENSDART00000162657 |
rps3
|
ribosomal protein S3 |
| chr20_-_40755614 | 5.26 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr10_+_6884627 | 5.20 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr14_-_12307522 | 5.19 |
ENSDART00000163900
|
myot
|
myotilin |
| chr15_-_20939579 | 5.19 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr16_-_29387215 | 5.11 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr21_+_25777425 | 5.09 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr6_+_50381665 | 5.07 |
ENSDART00000141128
|
cyc1
|
cytochrome c-1 |
| chr10_-_25217347 | 4.99 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr23_-_32157865 | 4.89 |
ENSDART00000000876
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr10_+_6884123 | 4.80 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr20_-_23426339 | 4.80 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr6_-_50203682 | 4.74 |
ENSDART00000083999
ENSDART00000143050 |
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr13_+_35637048 | 4.64 |
ENSDART00000085037
|
thbs2a
|
thrombospondin 2a |
| chr19_-_42556086 | 4.59 |
ENSDART00000051731
|
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr24_-_40744672 | 4.54 |
ENSDART00000160672
|
CU633479.1
|
|
| chr12_-_14143344 | 4.51 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr16_+_27349585 | 4.50 |
ENSDART00000142573
|
nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr9_-_21918963 | 4.39 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr21_-_5205617 | 4.38 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr13_+_25486608 | 4.37 |
ENSDART00000057689
|
bag3
|
BCL2 associated athanogene 3 |
| chr22_+_20427170 | 4.33 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr25_+_11456696 | 4.33 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr2_-_38284648 | 4.33 |
ENSDART00000148281
ENSDART00000132621 |
si:ch211-14a17.7
|
si:ch211-14a17.7 |
| chr5_+_37903790 | 4.32 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr20_+_20726231 | 4.30 |
ENSDART00000147112
|
zgc:193541
|
zgc:193541 |
| chr23_-_32162810 | 4.30 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr5_-_68093169 | 4.20 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr5_+_57924611 | 4.18 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr21_-_11654422 | 4.18 |
ENSDART00000081614
ENSDART00000132699 |
cast
|
calpastatin |
| chr20_+_52554352 | 4.13 |
ENSDART00000153217
ENSDART00000145230 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr23_+_36122058 | 4.10 |
ENSDART00000184448
|
hoxc3a
|
homeobox C3a |
| chr3_+_18398876 | 4.06 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr18_-_19456269 | 3.98 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr2_+_25198648 | 3.93 |
ENSDART00000110922
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr20_+_4060839 | 3.92 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr14_+_52565660 | 3.91 |
ENSDART00000188151
|
rpl26
|
ribosomal protein L26 |
| chr14_+_36521005 | 3.87 |
ENSDART00000192286
|
TENM3
|
si:dkey-237h12.3 |
| chr3_+_17537352 | 3.87 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr18_+_20560442 | 3.86 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr14_-_2933185 | 3.84 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr15_+_31344472 | 3.81 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr24_+_12835935 | 3.76 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr17_+_23462972 | 3.56 |
ENSDART00000112959
ENSDART00000192168 |
ankrd1a
|
ankyrin repeat domain 1a (cardiac muscle) |
| chr16_-_46579936 | 3.53 |
ENSDART00000166143
ENSDART00000127212 |
si:dkey-152b24.6
|
si:dkey-152b24.6 |
| chr8_+_41037541 | 3.51 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr9_+_29548195 | 3.51 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr3_-_30685401 | 3.43 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr3_-_29899172 | 3.40 |
ENSDART00000140518
ENSDART00000020311 |
rpl27
|
ribosomal protein L27 |
| chr18_-_16801033 | 3.39 |
ENSDART00000100100
|
admb
|
adrenomedullin b |
| chr13_+_38817871 | 3.37 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr14_+_34492288 | 3.36 |
ENSDART00000144301
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr24_-_3783497 | 3.33 |
ENSDART00000158354
|
adarb2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr5_-_9216758 | 3.33 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr1_+_6172786 | 3.31 |
ENSDART00000126468
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr2_-_30668580 | 3.31 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr22_-_35330532 | 3.30 |
ENSDART00000172654
|
FO818743.1
|
|
| chr13_-_50108337 | 3.30 |
ENSDART00000133308
|
nid1a
|
nidogen 1a |
| chr17_+_46818521 | 3.29 |
ENSDART00000156022
|
pimr14
|
Pim proto-oncogene, serine/threonine kinase, related 14 |
| chr16_+_32736588 | 3.29 |
ENSDART00000075191
ENSDART00000168358 |
zgc:172323
|
zgc:172323 |
| chr11_-_22303678 | 3.25 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr22_-_14161309 | 3.25 |
ENSDART00000133365
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr9_-_50001606 | 3.24 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr19_-_657439 | 3.23 |
ENSDART00000167100
|
slc6a18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr2_-_37098785 | 3.21 |
ENSDART00000003670
|
zgc:101744
|
zgc:101744 |
| chr18_+_23218980 | 3.19 |
ENSDART00000185014
|
mef2aa
|
myocyte enhancer factor 2aa |
| chr10_+_16584382 | 3.19 |
ENSDART00000112039
|
CR790388.1
|
|
| chr1_+_24387659 | 3.18 |
ENSDART00000130356
|
qdprb2
|
quinoid dihydropteridine reductase b2 |
| chr1_-_18811517 | 3.18 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr7_-_38792543 | 3.14 |
ENSDART00000157416
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr14_-_41075262 | 3.11 |
ENSDART00000180518
|
drp2
|
dystrophin related protein 2 |
| chr22_-_10459880 | 3.11 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr4_-_68563862 | 3.08 |
ENSDART00000182970
|
BX548011.2
|
|
| chr16_+_26777473 | 3.08 |
ENSDART00000188870
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr19_-_20403845 | 3.07 |
ENSDART00000151265
ENSDART00000147911 ENSDART00000151356 |
dazl
|
deleted in azoospermia-like |
| chr25_+_3549584 | 3.07 |
ENSDART00000165913
|
ccdc77
|
coiled-coil domain containing 77 |
| chr17_+_52612866 | 3.06 |
ENSDART00000182828
ENSDART00000191156 ENSDART00000188814 ENSDART00000109891 |
angel1
|
angel homolog 1 (Drosophila) |
| chr20_+_42978499 | 2.99 |
ENSDART00000138793
|
si:ch211-203k16.3
|
si:ch211-203k16.3 |
| chr24_+_1023839 | 2.97 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr18_+_15644559 | 2.96 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr14_+_44794936 | 2.95 |
ENSDART00000128881
|
zgc:195212
|
zgc:195212 |
| chr2_-_51794472 | 2.95 |
ENSDART00000186652
|
BX908782.3
|
|
| chr10_+_518546 | 2.90 |
ENSDART00000128275
|
npffr1l3
|
neuropeptide FF receptor 1 like 3 |
| chr20_-_38617766 | 2.89 |
ENSDART00000050474
|
slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr7_+_27317174 | 2.89 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr1_-_38195012 | 2.88 |
ENSDART00000020409
|
hand2
|
heart and neural crest derivatives expressed 2 |
| chr4_+_14660769 | 2.84 |
ENSDART00000168152
ENSDART00000013990 ENSDART00000079987 |
abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr13_+_2625150 | 2.84 |
ENSDART00000164177
|
plpp4
|
phospholipid phosphatase 4 |
| chr19_-_14191592 | 2.83 |
ENSDART00000164594
|
tbxta
|
T-box transcription factor Ta |
| chr23_+_404975 | 2.76 |
ENSDART00000181336
|
CU929146.1
|
|
| chr2_+_37227011 | 2.75 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr18_+_27821856 | 2.73 |
ENSDART00000131712
|
si:ch211-222m18.4
|
si:ch211-222m18.4 |
| chr1_-_29139141 | 2.68 |
ENSDART00000075546
ENSDART00000133246 |
hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr12_+_48803098 | 2.67 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr25_-_27621268 | 2.64 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr23_-_39666519 | 2.63 |
ENSDART00000110868
ENSDART00000190961 |
vwa1
|
von Willebrand factor A domain containing 1 |
| chr15_+_36309070 | 2.62 |
ENSDART00000157034
|
gmnc
|
geminin coiled-coil domain containing |
| chr12_+_2522642 | 2.62 |
ENSDART00000152567
|
frmpd2
|
FERM and PDZ domain containing 2 |
| chr1_-_513762 | 2.58 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr17_+_21295132 | 2.57 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr10_-_7756865 | 2.53 |
ENSDART00000114373
ENSDART00000125407 ENSDART00000016317 |
loxa
|
lysyl oxidase a |
| chr15_-_23376541 | 2.52 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr12_-_30359498 | 2.51 |
ENSDART00000152981
ENSDART00000189988 |
tdrd1
|
tudor domain containing 1 |
| chr13_-_12602920 | 2.51 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr14_+_45406299 | 2.51 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr15_-_4528326 | 2.50 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr15_-_2493771 | 2.49 |
ENSDART00000184906
|
neu4
|
sialidase 4 |
| chr14_-_29906209 | 2.49 |
ENSDART00000192952
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr24_+_2495197 | 2.49 |
ENSDART00000146887
|
f13a1a.1
|
coagulation factor XIII, A1 polypeptide a, tandem duplicate 1 |
| chr20_-_188461 | 2.45 |
ENSDART00000135530
|
smim8
|
small integral membrane protein 8 |
| chr8_+_37527575 | 2.45 |
ENSDART00000147239
|
or135-1
|
odorant receptor, family H, subfamily 135, member 1 |
| chr20_-_2619316 | 2.45 |
ENSDART00000185777
|
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr6_+_50381347 | 2.45 |
ENSDART00000055504
|
cyc1
|
cytochrome c-1 |
| chr8_+_26565512 | 2.44 |
ENSDART00000140980
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr4_-_77557279 | 2.43 |
ENSDART00000180113
|
AL935186.10
|
|
| chr1_-_55248496 | 2.40 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr24_+_2470061 | 2.39 |
ENSDART00000140383
ENSDART00000191261 |
F13A1 (1 of many)
|
coagulation factor XIII A chain |
| chr9_+_29548630 | 2.37 |
ENSDART00000132295
|
rnf17
|
ring finger protein 17 |
| chr9_-_19161982 | 2.35 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr8_+_18044257 | 2.35 |
ENSDART00000029255
|
glis1b
|
GLIS family zinc finger 1b |
| chr19_-_20403507 | 2.35 |
ENSDART00000052603
ENSDART00000137590 |
dazl
|
deleted in azoospermia-like |
| chr12_-_30358836 | 2.32 |
ENSDART00000152878
|
tdrd1
|
tudor domain containing 1 |
| chr16_-_16761164 | 2.32 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr11_+_7214353 | 2.31 |
ENSDART00000156764
|
nwd1
|
NACHT and WD repeat domain containing 1 |
| chr6_+_52873822 | 2.31 |
ENSDART00000103138
|
or137-3
|
odorant receptor, family H, subfamily 137, member 3 |
| chr25_+_13620555 | 2.31 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr12_-_30359031 | 2.30 |
ENSDART00000192628
|
tdrd1
|
tudor domain containing 1 |
| chr24_+_5912635 | 2.30 |
ENSDART00000153736
|
pimr63
|
Pim proto-oncogene, serine/threonine kinase, related 63 |
| chr6_+_41191482 | 2.30 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr15_-_21877726 | 2.29 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr17_-_48944465 | 2.29 |
ENSDART00000154110
|
si:ch1073-80i24.3
|
si:ch1073-80i24.3 |
| chr15_+_34988148 | 2.26 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr15_-_36727462 | 2.25 |
ENSDART00000085971
|
nphs1
|
nephrosis 1, congenital, Finnish type (nephrin) |
| chr21_-_18993110 | 2.23 |
ENSDART00000144086
|
si:ch211-222n4.6
|
si:ch211-222n4.6 |
| chr17_-_37395460 | 2.22 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr18_-_33979422 | 2.22 |
ENSDART00000136535
ENSDART00000167698 |
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr4_+_9679840 | 2.22 |
ENSDART00000190889
|
BX901962.12
|
|
| chr23_+_24272421 | 2.22 |
ENSDART00000029974
|
clcnk
|
chloride channel K |
| chr18_+_32801603 | 2.20 |
ENSDART00000163462
|
olfcg6
|
olfactory receptor C family, g6 |
| chr4_-_12907826 | 2.20 |
ENSDART00000182923
|
msrb3
|
methionine sulfoxide reductase B3 |
| chr16_+_30483043 | 2.18 |
ENSDART00000188034
|
pear1
|
platelet endothelial aggregation receptor 1 |
| chr23_-_17003533 | 2.17 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr14_+_50034196 | 2.16 |
ENSDART00000190301
|
LO018202.1
|
|
| chr16_-_42056137 | 2.16 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr7_+_1467863 | 2.15 |
ENSDART00000173433
|
emc4
|
ER membrane protein complex subunit 4 |
| chr21_-_31290582 | 2.14 |
ENSDART00000065362
|
ca4c
|
carbonic anhydrase IV c |
| chr13_+_28701233 | 2.14 |
ENSDART00000135931
|
si:ch211-67n3.9
|
si:ch211-67n3.9 |
| chr14_+_34490445 | 2.14 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr25_+_3035384 | 2.13 |
ENSDART00000184219
ENSDART00000149360 |
mpi
|
mannose phosphate isomerase |
| chr1_+_56873359 | 2.13 |
ENSDART00000152713
|
si:ch211-152f2.2
|
si:ch211-152f2.2 |
| chr11_+_12052791 | 2.13 |
ENSDART00000158479
|
si:ch211-156l18.8
|
si:ch211-156l18.8 |
| chr3_+_12816362 | 2.12 |
ENSDART00000163743
ENSDART00000170788 |
cyp2k6
|
cytochrome P450, family 2, subfamily K, polypeptide 6 |
| chr23_-_36823932 | 2.12 |
ENSDART00000142305
|
hipk1a
|
homeodomain interacting protein kinase 1a |
| chr8_-_46525092 | 2.11 |
ENSDART00000030482
|
sult1st2
|
sulfotransferase family 1, cytosolic sulfotransferase 2 |
| chr17_-_23416897 | 2.11 |
ENSDART00000163391
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr6_+_16736871 | 2.11 |
ENSDART00000155471
|
pimr12
|
Pim proto-oncogene, serine/threonine kinase, related 12 |
| chr24_+_17269849 | 2.11 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr1_-_55068941 | 2.11 |
ENSDART00000152143
ENSDART00000152590 |
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr23_+_39695827 | 2.10 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr10_+_11261576 | 2.10 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr1_-_21287724 | 2.10 |
ENSDART00000193900
|
npy1r
|
neuropeptide Y receptor Y1 |
| chr16_-_32304764 | 2.09 |
ENSDART00000143859
ENSDART00000134381 |
mms22l
|
MMS22-like, DNA repair protein |
| chr18_+_33570170 | 2.09 |
ENSDART00000133276
|
si:dkey-47k20.3
|
si:dkey-47k20.3 |
| chr5_+_6954162 | 2.09 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr11_-_1550709 | 2.08 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr14_-_8940499 | 2.08 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
Network of associatons between targets according to the STRING database.
First level regulatory network of alx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 22.8 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 2.4 | 2.4 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 1.8 | 5.5 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 1.6 | 6.3 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 1.5 | 13.7 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 1.5 | 78.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 1.5 | 4.4 | GO:0010664 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 1.3 | 3.9 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 1.3 | 3.8 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 1.2 | 1.2 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) sarcoplasmic reticulum calcium ion transport(GO:0070296) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 1.0 | 5.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 1.0 | 3.0 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 1.0 | 3.9 | GO:0042755 | eating behavior(GO:0042755) |
| 1.0 | 12.4 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.9 | 9.2 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.9 | 5.4 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.9 | 3.5 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.8 | 4.2 | GO:0015743 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.8 | 7.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.7 | 2.2 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.7 | 8.1 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.7 | 10.0 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.7 | 2.8 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.7 | 2.1 | GO:0046222 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.7 | 2.0 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.6 | 11.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.6 | 0.6 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.6 | 4.2 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.6 | 6.4 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.6 | 2.9 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.6 | 2.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.5 | 4.9 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.5 | 2.6 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.5 | 2.6 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.5 | 8.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.5 | 1.5 | GO:2000376 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.5 | 2.9 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.5 | 1.4 | GO:0045887 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.5 | 2.3 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.4 | 1.3 | GO:0009595 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.4 | 2.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.4 | 3.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.4 | 2.1 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.4 | 1.6 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.4 | 1.5 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.4 | 1.8 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.3 | 2.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.3 | 7.6 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.3 | 4.0 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.3 | 2.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.3 | 2.2 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.3 | 2.0 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.3 | 2.9 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.3 | 2.0 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.3 | 2.8 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.3 | 1.1 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.3 | 3.2 | GO:0006559 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.2 | 2.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.2 | 4.4 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.2 | 2.7 | GO:0097300 | necrotic cell death(GO:0070265) cellular response to hydrogen peroxide(GO:0070301) programmed necrotic cell death(GO:0097300) |
| 0.2 | 7.7 | GO:0045761 | regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.2 | 1.6 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.2 | 2.1 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.2 | 1.4 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.2 | 3.6 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.2 | 2.0 | GO:0060114 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.2 | 1.3 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.2 | 2.5 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.2 | 1.5 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.2 | 2.0 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.2 | 4.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 2.5 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.2 | 1.5 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.2 | 6.2 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.2 | 3.0 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.2 | 1.8 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.2 | 1.1 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.2 | 2.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 1.9 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.2 | 2.5 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 7.9 | GO:0018198 | peptidyl-cysteine modification(GO:0018198) |
| 0.2 | 1.9 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.2 | 0.5 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.2 | 1.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.2 | 0.6 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.2 | 3.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 0.4 | GO:0061037 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.1 | 2.0 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 1.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 1.2 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.1 | 1.5 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 1.7 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 1.2 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 14.2 | GO:0001933 | negative regulation of protein phosphorylation(GO:0001933) |
| 0.1 | 1.0 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 1.9 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.1 | 1.5 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 3.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 2.5 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 1.9 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 2.0 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 1.4 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 0.6 | GO:0046084 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.1 | 2.8 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 0.3 | GO:1902767 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.1 | 0.8 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 1.8 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 2.9 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.8 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.0 | GO:0019430 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 0.7 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 1.6 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.1 | 3.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 2.1 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.1 | 0.6 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 8.0 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.1 | 0.3 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 1.7 | GO:0070672 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 2.1 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 1.2 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 1.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 4.9 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 5.0 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.1 | 1.6 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.7 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 1.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 4.4 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.9 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.1 | 0.5 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 1.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.3 | GO:0045598 | regulation of fat cell differentiation(GO:0045598) |
| 0.1 | 3.0 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 2.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 2.2 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 1.0 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.0 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 3.9 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 2.2 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 0.3 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.1 | 1.2 | GO:0051784 | negative regulation of mitotic nuclear division(GO:0045839) negative regulation of nuclear division(GO:0051784) |
| 0.1 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.9 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.3 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 4.1 | GO:1903522 | regulation of blood circulation(GO:1903522) |
| 0.1 | 2.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 1.0 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 1.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.4 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 2.2 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 1.1 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 2.1 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.9 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 1.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:0060343 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.0 | 1.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.9 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) |
| 0.0 | 0.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.0 | 2.7 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.7 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.2 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 3.8 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.3 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.7 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 3.3 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 0.5 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 1.4 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.7 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 1.4 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 3.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 4.2 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 2.0 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 8.1 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 9.9 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.4 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 1.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.6 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 2.8 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 1.7 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.2 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) regulation of T cell migration(GO:2000404) |
| 0.0 | 0.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 1.5 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.4 | GO:0060047 | heart contraction(GO:0060047) |
| 0.0 | 0.2 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 6.8 | GO:0043043 | peptide biosynthetic process(GO:0043043) |
| 0.0 | 0.3 | GO:0048515 | spermatid development(GO:0007286) spermatid differentiation(GO:0048515) |
| 0.0 | 2.3 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.5 | GO:0032835 | glomerulus development(GO:0032835) |
| 0.0 | 0.5 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 2.0 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 13.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 1.8 | 5.5 | GO:0097189 | apoptotic body(GO:0097189) |
| 1.8 | 78.3 | GO:0005861 | troponin complex(GO:0005861) |
| 1.1 | 3.4 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 1.1 | 8.6 | GO:0071546 | pi-body(GO:0071546) |
| 0.8 | 7.5 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.7 | 8.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.7 | 5.8 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.6 | 4.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.5 | 13.0 | GO:0043186 | P granule(GO:0043186) |
| 0.3 | 2.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.3 | 2.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 3.8 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.3 | 4.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 15.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.3 | 28.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.3 | 3.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 2.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 1.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 1.2 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.2 | 2.5 | GO:0035101 | FACT complex(GO:0035101) |
| 0.2 | 1.8 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.2 | 4.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 9.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 2.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 2.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 5.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 2.8 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.1 | 1.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 1.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 5.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.6 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 2.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.7 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 2.7 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.4 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 1.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 4.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.6 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 2.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 19.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.6 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 4.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 13.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 3.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 4.8 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 2.6 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 2.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.0 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.4 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 10.6 | GO:0044429 | mitochondrial part(GO:0044429) |
| 0.0 | 0.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 1.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.1 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.0 | 19.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 7.3 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 4.1 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 2.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 15.8 | GO:0009374 | biotin binding(GO:0009374) |
| 2.2 | 6.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 1.7 | 6.7 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.2 | 13.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 1.0 | 12.4 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.9 | 6.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.9 | 7.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.9 | 3.6 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.8 | 4.2 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.8 | 3.2 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.8 | 11.3 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.7 | 8.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.7 | 3.3 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.6 | 2.6 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.6 | 5.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.6 | 4.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.6 | 3.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.6 | 3.5 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.6 | 6.4 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.6 | 2.8 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.5 | 1.6 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.5 | 8.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.5 | 3.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.5 | 2.5 | GO:0016415 | octanoyltransferase activity(GO:0016415) |
| 0.5 | 1.5 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.5 | 1.9 | GO:0004945 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.5 | 2.3 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.4 | 1.3 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.4 | 3.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.4 | 4.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 6.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.4 | 2.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.3 | 2.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 2.0 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.3 | 1.6 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.3 | 10.4 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.3 | 1.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 0.7 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 0.7 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.2 | 2.0 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.2 | 1.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 2.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.2 | 4.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 2.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 1.8 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.2 | 2.5 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.2 | 1.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 2.9 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 0.7 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 1.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 3.7 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.2 | 10.4 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.2 | 5.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 2.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.2 | 3.0 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.2 | 1.4 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 2.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 1.3 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 7.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 1.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 4.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 2.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.0 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 1.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 1.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 16.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 2.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 3.1 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 4.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 22.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 1.6 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.9 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 8.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.3 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 1.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.9 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.3 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.1 | 2.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 5.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.7 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 6.5 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.1 | 0.6 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.1 | 1.5 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.3 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 1.7 | GO:0005501 | retinoid binding(GO:0005501) isoprenoid binding(GO:0019840) |
| 0.1 | 2.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.9 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 10.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 1.7 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 2.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 2.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.6 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 2.9 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 3.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.7 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 0.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.5 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 4.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.6 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 3.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 2.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 2.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.1 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 1.0 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 13.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 2.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.3 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 1.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 1.0 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 7.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 2.0 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 4.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.1 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 2.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 2.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 6.8 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 7.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.9 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 3.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.0 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 12.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.3 | 9.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 4.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 9.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 2.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 18.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 3.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 3.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 4.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 2.8 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 1.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 22.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.5 | 3.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.5 | 10.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.5 | 3.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 13.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 3.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 2.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 5.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 1.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 25.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 3.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 2.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 2.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 1.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 6.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 6.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 0.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 4.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 4.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.9 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 2.6 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.9 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |