Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
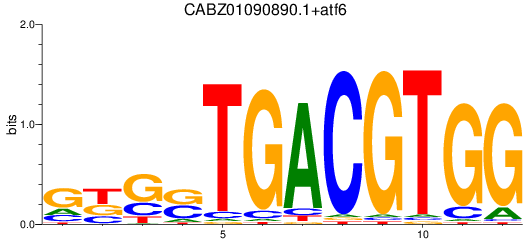
Results for CABZ01090890.1+atf6
Z-value: 1.35
Transcription factors associated with CABZ01090890.1+atf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
atf6
|
ENSDARG00000012656 | activating transcription factor 6 |
|
CABZ01090890.1
|
ENSDARG00000101369 | ENSDARG00000101369 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| atf6 | dr11_v1_chr20_-_33864804_33864804 | 0.83 | 3.9e-25 | Click! |
Activity profile of CABZ01090890.1+atf6 motif
Sorted Z-values of CABZ01090890.1+atf6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_33606739 | 11.41 |
ENSDART00000026464
|
cfl1l
|
cofilin 1 (non-muscle), like |
| chr16_-_45178430 | 8.43 |
ENSDART00000165186
|
si:dkey-33i11.9
|
si:dkey-33i11.9 |
| chr5_-_9540641 | 7.52 |
ENSDART00000124384
ENSDART00000160079 |
gak
|
cyclin G associated kinase |
| chr6_+_13506841 | 6.86 |
ENSDART00000032331
|
gmppab
|
GDP-mannose pyrophosphorylase Ab |
| chr22_+_1796057 | 6.82 |
ENSDART00000170834
|
znf1179
|
zinc finger protein 1179 |
| chr12_-_17479078 | 6.21 |
ENSDART00000079115
|
papss2b
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2b |
| chr21_+_11105007 | 6.19 |
ENSDART00000128859
ENSDART00000193105 |
prlra
|
prolactin receptor a |
| chr14_-_11456724 | 5.95 |
ENSDART00000110424
|
si:ch211-153b23.4
|
si:ch211-153b23.4 |
| chr17_-_22324727 | 5.46 |
ENSDART00000160341
|
CU104709.1
|
|
| chr18_+_21122818 | 5.39 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr21_-_26677834 | 5.35 |
ENSDART00000077381
|
nxf1
|
nuclear RNA export factor 1 |
| chr21_+_5915041 | 5.27 |
ENSDART00000151370
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr2_+_10063747 | 5.25 |
ENSDART00000143876
ENSDART00000014088 ENSDART00000134554 |
cmpk
|
cytidylate kinase |
| chr11_+_42494531 | 5.18 |
ENSDART00000067604
|
arf4a
|
ADP-ribosylation factor 4a |
| chr6_-_607063 | 5.01 |
ENSDART00000189900
|
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr20_-_40717900 | 4.99 |
ENSDART00000181663
|
cx43
|
connexin 43 |
| chr16_-_52540056 | 4.96 |
ENSDART00000188304
|
CR293507.1
|
|
| chr23_-_31266586 | 4.85 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr3_-_18384501 | 4.71 |
ENSDART00000027630
|
kdelr2a
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2a |
| chr19_-_30404096 | 4.53 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr21_+_10702031 | 4.48 |
ENSDART00000102304
|
lman1
|
lectin, mannose-binding, 1 |
| chr4_+_47257854 | 4.37 |
ENSDART00000173868
|
crestin
|
crestin |
| chr9_+_24192370 | 4.31 |
ENSDART00000003482
|
stk17b
|
serine/threonine kinase 17b (apoptosis-inducing) |
| chr21_-_25295087 | 4.31 |
ENSDART00000087910
ENSDART00000147860 |
st14b
|
suppression of tumorigenicity 14 (colon carcinoma) b |
| chr23_-_25798099 | 4.21 |
ENSDART00000041833
|
fitm2
|
fat storage-inducing transmembrane protein 2 |
| chr4_-_991043 | 4.20 |
ENSDART00000184706
|
naga
|
N-acetylgalactosaminidase, alpha |
| chr1_+_38142715 | 4.11 |
ENSDART00000079928
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr1_+_59146298 | 4.10 |
ENSDART00000191885
ENSDART00000152747 |
gpr108
|
G protein-coupled receptor 108 |
| chr21_+_10701834 | 4.03 |
ENSDART00000192473
|
lman1
|
lectin, mannose-binding, 1 |
| chr5_-_1487256 | 3.99 |
ENSDART00000149599
ENSDART00000148411 ENSDART00000092087 ENSDART00000148464 |
golga2
|
golgin A2 |
| chr2_-_3045861 | 3.97 |
ENSDART00000105818
ENSDART00000187575 |
guk1a
|
guanylate kinase 1a |
| chr16_-_54942532 | 3.96 |
ENSDART00000078887
ENSDART00000101402 |
tmem222a
|
transmembrane protein 222a |
| chr19_-_30403922 | 3.89 |
ENSDART00000181841
|
agr2
|
anterior gradient 2 |
| chr14_+_23687678 | 3.88 |
ENSDART00000002469
|
hspa4b
|
heat shock protein 4b |
| chr4_-_13921185 | 3.81 |
ENSDART00000143202
ENSDART00000080334 |
yaf2
|
YY1 associated factor 2 |
| chr21_+_30355767 | 3.69 |
ENSDART00000189948
|
CR749164.1
|
|
| chr2_-_11504347 | 3.66 |
ENSDART00000019392
|
sdr16c5a
|
short chain dehydrogenase/reductase family 16C, member 5a |
| chr3_-_46410387 | 3.60 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr17_-_49438873 | 3.60 |
ENSDART00000004424
|
znf292a
|
zinc finger protein 292a |
| chr1_+_26605065 | 3.59 |
ENSDART00000011645
|
coro2a
|
coronin, actin binding protein, 2A |
| chr10_+_5689510 | 3.54 |
ENSDART00000183217
ENSDART00000172632 |
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr3_+_36616713 | 3.49 |
ENSDART00000158284
|
pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr22_+_32228882 | 3.44 |
ENSDART00000092082
|
manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr5_-_58939460 | 3.42 |
ENSDART00000122413
|
mcama
|
melanoma cell adhesion molecule a |
| chr25_-_3034668 | 3.41 |
ENSDART00000053405
|
scamp2
|
secretory carrier membrane protein 2 |
| chr13_-_45123051 | 3.39 |
ENSDART00000138644
|
runx3
|
runt-related transcription factor 3 |
| chr19_+_17400283 | 3.38 |
ENSDART00000127353
|
nr1d2b
|
nuclear receptor subfamily 1, group D, member 2b |
| chr20_+_26940178 | 3.34 |
ENSDART00000190888
|
cdca4
|
cell division cycle associated 4 |
| chr10_+_42374957 | 3.34 |
ENSDART00000147926
|
COX5B
|
zgc:86599 |
| chr15_-_25153352 | 3.32 |
ENSDART00000078095
ENSDART00000122184 |
vps53
|
vacuolar protein sorting 53 homolog (S. cerevisiae) |
| chr15_+_3808996 | 3.32 |
ENSDART00000110227
|
RNF14 (1 of many)
|
ring finger protein 14 |
| chr4_-_25515513 | 3.31 |
ENSDART00000142276
ENSDART00000044043 |
rbm17
|
RNA binding motif protein 17 |
| chr19_+_31904836 | 3.28 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr3_-_55537096 | 3.24 |
ENSDART00000123544
ENSDART00000188752 |
tex2
|
testis expressed 2 |
| chr17_-_31344400 | 3.23 |
ENSDART00000154013
|
bahd1
|
bromo adjacent homology domain containing 1 |
| chr3_-_57762247 | 3.23 |
ENSDART00000156522
|
cant1a
|
calcium activated nucleotidase 1a |
| chr10_+_22510280 | 3.20 |
ENSDART00000109070
ENSDART00000182002 ENSDART00000192852 |
gigyf1b
|
GRB10 interacting GYF protein 1b |
| chr22_+_10713713 | 3.17 |
ENSDART00000122349
|
hiat1b
|
hippocampus abundant transcript 1b |
| chr8_-_44298964 | 3.16 |
ENSDART00000098520
|
fzd10
|
frizzled class receptor 10 |
| chr3_+_36617024 | 3.14 |
ENSDART00000189957
|
pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr15_+_47746176 | 3.13 |
ENSDART00000154481
ENSDART00000160914 |
stard10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr4_+_357810 | 3.10 |
ENSDART00000163436
ENSDART00000103645 |
tmem181
|
transmembrane protein 181 |
| chr17_-_28797395 | 3.09 |
ENSDART00000134735
|
scfd1
|
sec1 family domain containing 1 |
| chr11_+_440305 | 3.05 |
ENSDART00000082517
|
rab43
|
RAB43, member RAS oncogene family |
| chr17_-_14705039 | 3.04 |
ENSDART00000154281
ENSDART00000123550 |
ptp4a2a
|
protein tyrosine phosphatase type IVA, member 2a |
| chr3_-_47235997 | 3.03 |
ENSDART00000047071
|
tmed1a
|
transmembrane p24 trafficking protein 1a |
| chr10_+_45148167 | 3.02 |
ENSDART00000172621
|
ogdhb
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase b (lipoamide) |
| chr9_-_42418470 | 3.00 |
ENSDART00000144353
|
calcrla
|
calcitonin receptor-like a |
| chr23_+_20431140 | 2.93 |
ENSDART00000193950
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr10_-_8672820 | 2.85 |
ENSDART00000080763
|
si:dkey-27b3.2
|
si:dkey-27b3.2 |
| chr4_+_13568469 | 2.83 |
ENSDART00000171235
ENSDART00000136152 |
calua
|
calumenin a |
| chr11_-_22916641 | 2.81 |
ENSDART00000080201
ENSDART00000154813 |
mdm4
|
MDM4, p53 regulator |
| chr13_-_23665580 | 2.80 |
ENSDART00000144282
|
map3k21
|
mitogen-activated protein kinase kinase kinase 21 |
| chr4_-_27099224 | 2.80 |
ENSDART00000048383
|
creld2
|
cysteine-rich with EGF-like domains 2 |
| chr19_+_10592778 | 2.78 |
ENSDART00000135488
ENSDART00000151624 |
si:dkey-211g8.5
|
si:dkey-211g8.5 |
| chr3_-_36750068 | 2.76 |
ENSDART00000173388
|
abcc6b.1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6b, tandem duplicate 1 |
| chr17_+_21102301 | 2.75 |
ENSDART00000035432
|
entpd6
|
ectonucleoside triphosphate diphosphohydrolase 6 (putative) |
| chr15_-_43978141 | 2.74 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr10_+_16036573 | 2.73 |
ENSDART00000188757
|
lmnb1
|
lamin B1 |
| chr10_-_15879569 | 2.72 |
ENSDART00000136789
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr18_-_46280578 | 2.67 |
ENSDART00000131724
|
pld3
|
phospholipase D family, member 3 |
| chr1_+_55080797 | 2.64 |
ENSDART00000077390
|
lgalsla
|
lectin, galactoside-binding-like a |
| chr6_-_38930726 | 2.59 |
ENSDART00000154151
|
hdac7b
|
histone deacetylase 7b |
| chr3_+_22984098 | 2.58 |
ENSDART00000043190
|
lsm12a
|
LSM12 homolog a |
| chr15_-_163586 | 2.57 |
ENSDART00000163597
|
SEPT4
|
septin-4 |
| chr4_+_23117557 | 2.56 |
ENSDART00000066909
|
slc35e3
|
solute carrier family 35, member E3 |
| chr19_-_10324573 | 2.55 |
ENSDART00000171795
|
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
| chr1_+_36771954 | 2.52 |
ENSDART00000149022
|
arhgap10
|
Rho GTPase activating protein 10 |
| chr15_+_41815703 | 2.51 |
ENSDART00000059508
|
pxylp1
|
2-phosphoxylose phosphatase 1 |
| chr14_-_45595711 | 2.48 |
ENSDART00000074038
|
scyl1
|
SCY1-like, kinase-like 1 |
| chr18_-_34549721 | 2.48 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr25_-_37084032 | 2.46 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr21_-_2124497 | 2.42 |
ENSDART00000166003
|
si:rp71-1h20.5
|
si:rp71-1h20.5 |
| chr25_-_17378881 | 2.40 |
ENSDART00000064586
|
cyp2x7
|
cytochrome P450, family 2, subfamily X, polypeptide 7 |
| chr3_-_26183699 | 2.39 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr17_+_10501647 | 2.38 |
ENSDART00000140391
|
tyro3
|
TYRO3 protein tyrosine kinase |
| chr6_-_28943056 | 2.38 |
ENSDART00000065138
|
tbc1d23
|
TBC1 domain family, member 23 |
| chr20_-_51727860 | 2.31 |
ENSDART00000147044
|
brox
|
BRO1 domain and CAAX motif containing |
| chr23_+_7692042 | 2.29 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr5_-_48680580 | 2.29 |
ENSDART00000031194
|
lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr7_+_19835569 | 2.29 |
ENSDART00000149812
|
ovol1a
|
ovo-like zinc finger 1a |
| chr3_-_22829710 | 2.27 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr9_+_21793565 | 2.24 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr4_-_25515154 | 2.21 |
ENSDART00000186524
|
rbm17
|
RNA binding motif protein 17 |
| chr4_-_73488406 | 2.20 |
ENSDART00000115002
|
si:ch73-266f23.1
|
si:ch73-266f23.1 |
| chr6_-_1591002 | 2.20 |
ENSDART00000087039
|
zgc:123305
|
zgc:123305 |
| chr12_+_31783066 | 2.19 |
ENSDART00000105584
|
lrrc59
|
leucine rich repeat containing 59 |
| chr10_+_5060191 | 2.19 |
ENSDART00000145908
ENSDART00000122397 |
carm1l
|
coactivator-associated arginine methyltransferase 1, like |
| chr9_+_8898835 | 2.16 |
ENSDART00000147820
|
naxd
|
NAD(P)HX dehydratase |
| chr19_-_10324182 | 2.15 |
ENSDART00000151352
ENSDART00000151162 ENSDART00000023571 |
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
| chr7_-_6086732 | 2.15 |
ENSDART00000189160
|
si:cabz01036006.1
|
si:cabz01036006.1 |
| chr12_-_13318944 | 2.15 |
ENSDART00000152201
ENSDART00000041394 |
emc9
|
ER membrane protein complex subunit 9 |
| chr16_-_36064143 | 2.15 |
ENSDART00000158358
ENSDART00000182584 |
stk40
|
serine/threonine kinase 40 |
| chr8_-_5220125 | 2.12 |
ENSDART00000035676
|
bnip3la
|
BCL2 interacting protein 3 like a |
| chr6_-_50685862 | 2.12 |
ENSDART00000134146
|
mtss1
|
metastasis suppressor 1 |
| chr1_+_38142354 | 2.09 |
ENSDART00000179352
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr2_-_38225388 | 2.09 |
ENSDART00000146485
ENSDART00000128043 |
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr23_-_29668286 | 2.08 |
ENSDART00000129248
|
clstn1
|
calsyntenin 1 |
| chr22_+_2860260 | 2.05 |
ENSDART00000106221
|
si:dkey-20i20.2
|
si:dkey-20i20.2 |
| chr22_+_24157807 | 2.02 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr6_+_27275716 | 1.99 |
ENSDART00000156434
ENSDART00000114347 |
scly
|
selenocysteine lyase |
| chr7_-_69352424 | 1.98 |
ENSDART00000170714
|
ap1g1
|
adaptor-related protein complex 1, gamma 1 subunit |
| chr4_-_8030583 | 1.97 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr14_-_4044545 | 1.91 |
ENSDART00000169527
|
snx25
|
sorting nexin 25 |
| chr6_+_55428924 | 1.91 |
ENSDART00000018270
|
ncoa5
|
nuclear receptor coactivator 5 |
| chr1_-_46663997 | 1.90 |
ENSDART00000134450
|
ebpl
|
emopamil binding protein-like |
| chr17_+_10090638 | 1.89 |
ENSDART00000169522
ENSDART00000160156 |
sec23a
|
Sec23 homolog A, COPII coat complex component |
| chr21_+_19858627 | 1.87 |
ENSDART00000147010
|
fybb
|
FYN binding protein b |
| chr9_-_32672129 | 1.87 |
ENSDART00000140581
|
gzm3.4
|
granzyme 3, tandem duplicate 4 |
| chr9_+_54237100 | 1.85 |
ENSDART00000148928
|
rbm27
|
RNA binding motif protein 27 |
| chr4_+_12931763 | 1.85 |
ENSDART00000016382
|
wif1
|
wnt inhibitory factor 1 |
| chr6_+_58289335 | 1.85 |
ENSDART00000177399
|
ralgapb
|
Ral GTPase activating protein, beta subunit (non-catalytic) |
| chr11_+_42482920 | 1.83 |
ENSDART00000160937
|
arf4a
|
ADP-ribosylation factor 4a |
| chr7_+_13995792 | 1.83 |
ENSDART00000091470
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr1_+_10018466 | 1.82 |
ENSDART00000113551
|
trim2b
|
tripartite motif containing 2b |
| chr3_-_31845816 | 1.81 |
ENSDART00000157028
|
map3k3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr24_-_6158933 | 1.81 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr11_-_42134968 | 1.80 |
ENSDART00000187115
|
FP325130.1
|
|
| chr23_+_33718602 | 1.79 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr8_-_31919624 | 1.79 |
ENSDART00000085573
|
rgs7bpa
|
regulator of G protein signaling 7 binding protein a |
| chr1_+_8662530 | 1.73 |
ENSDART00000054989
|
fscn1b
|
fascin actin-bundling protein 1b |
| chr22_+_1947494 | 1.71 |
ENSDART00000159121
|
si:dkey-15h8.15
|
si:dkey-15h8.15 |
| chr14_-_12390724 | 1.66 |
ENSDART00000131343
|
magt1
|
magnesium transporter 1 |
| chr3_-_26184018 | 1.65 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr18_-_26894732 | 1.63 |
ENSDART00000147735
ENSDART00000188938 |
BX470164.1
si:dkey-24l11.2
|
si:dkey-24l11.2 |
| chr21_+_19648814 | 1.61 |
ENSDART00000048581
|
fgf10a
|
fibroblast growth factor 10a |
| chr3_+_24094581 | 1.61 |
ENSDART00000138270
ENSDART00000131509 |
copz2
|
coatomer protein complex, subunit zeta 2 |
| chr6_+_59176470 | 1.59 |
ENSDART00000161720
|
shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr10_+_16036246 | 1.58 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr9_+_33261330 | 1.58 |
ENSDART00000135384
|
usp9
|
ubiquitin specific peptidase 9 |
| chr6_+_13742899 | 1.55 |
ENSDART00000104722
|
cdk5r2a
|
cyclin-dependent kinase 5, regulatory subunit 2a (p39) |
| chr23_+_2703044 | 1.54 |
ENSDART00000182512
ENSDART00000105286 |
ncoa6
|
nuclear receptor coactivator 6 |
| chr10_+_42374770 | 1.54 |
ENSDART00000020000
|
COX5B
|
zgc:86599 |
| chr15_+_17100697 | 1.54 |
ENSDART00000183565
ENSDART00000123197 |
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr13_-_25842074 | 1.51 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr7_+_39166460 | 1.50 |
ENSDART00000052318
ENSDART00000146635 ENSDART00000173877 ENSDART00000173767 ENSDART00000173600 |
mdka
|
midkine a |
| chr5_-_27994679 | 1.50 |
ENSDART00000132740
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr22_-_10641873 | 1.47 |
ENSDART00000064772
|
cyb561d2
|
cytochrome b561 family, member D2 |
| chr22_+_1786230 | 1.46 |
ENSDART00000169318
ENSDART00000164948 |
znf1154
|
zinc finger protein 1154 |
| chr15_+_24756860 | 1.46 |
ENSDART00000156424
ENSDART00000078035 |
cpda
|
carboxypeptidase D, a |
| chr15_-_47937204 | 1.44 |
ENSDART00000154705
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr21_-_40413191 | 1.44 |
ENSDART00000003221
|
nsrp1
|
nuclear speckle splicing regulatory protein 1 |
| chr9_+_22780901 | 1.41 |
ENSDART00000110992
ENSDART00000143972 |
rif1
|
replication timing regulatory factor 1 |
| chr11_-_3897067 | 1.40 |
ENSDART00000134858
|
rpn1
|
ribophorin I |
| chr18_-_25855263 | 1.40 |
ENSDART00000042074
|
sema4ba
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ba |
| chr22_+_2254972 | 1.40 |
ENSDART00000144906
|
znf1157
|
zinc finger protein 1157 |
| chr24_-_20652473 | 1.38 |
ENSDART00000179984
|
nktr
|
natural killer cell triggering receptor |
| chr9_-_8979154 | 1.35 |
ENSDART00000145266
|
ing1
|
inhibitor of growth family, member 1 |
| chr18_-_5248365 | 1.26 |
ENSDART00000082506
ENSDART00000082504 ENSDART00000097960 |
myef2
|
myelin expression factor 2 |
| chr4_-_3145359 | 1.24 |
ENSDART00000112210
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr23_+_20431388 | 1.24 |
ENSDART00000132920
ENSDART00000102963 ENSDART00000109899 ENSDART00000140219 |
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr4_+_17642731 | 1.23 |
ENSDART00000026509
|
cwf19l1
|
CWF19-like 1, cell cycle control |
| chr19_+_4443285 | 1.23 |
ENSDART00000162683
|
trappc9
|
trafficking protein particle complex 9 |
| chr15_-_35212462 | 1.23 |
ENSDART00000043960
|
agfg1a
|
ArfGAP with FG repeats 1a |
| chr6_-_29051773 | 1.20 |
ENSDART00000190508
ENSDART00000180191 ENSDART00000111682 |
evi5b
|
ecotropic viral integration site 5b |
| chr17_-_24879003 | 1.19 |
ENSDART00000123147
|
zbtb8a
|
zinc finger and BTB domain containing 8A |
| chr4_-_19693978 | 1.19 |
ENSDART00000100974
ENSDART00000040405 |
snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr8_+_1843135 | 1.16 |
ENSDART00000141452
|
snap29
|
synaptosomal-associated protein 29 |
| chr5_+_25585869 | 1.16 |
ENSDART00000138060
|
si:dkey-229d2.7
|
si:dkey-229d2.7 |
| chr13_+_36923052 | 1.13 |
ENSDART00000026313
|
tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr15_+_17100412 | 1.13 |
ENSDART00000154418
|
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr22_+_1853999 | 1.12 |
ENSDART00000163288
|
znf1174
|
zinc finger protein 1174 |
| chr1_+_10110203 | 1.09 |
ENSDART00000080576
ENSDART00000181437 |
lratb.1
|
lecithin retinol acyltransferase b, tandem duplicate 1 |
| chr22_+_2239254 | 1.07 |
ENSDART00000131396
ENSDART00000135320 |
znf1144
|
zinc finger protein 1144 |
| chr12_+_33919502 | 1.05 |
ENSDART00000085888
|
trim8b
|
tripartite motif containing 8b |
| chr19_-_7540821 | 1.04 |
ENSDART00000143958
|
lix1l
|
limb and CNS expressed 1 like |
| chr16_+_1228073 | 1.03 |
ENSDART00000109645
|
LO017852.1
|
|
| chr8_-_14052349 | 1.02 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr19_-_29294457 | 1.02 |
ENSDART00000130815
ENSDART00000103437 |
e2f3
|
E2F transcription factor 3 |
| chr2_+_49864219 | 1.02 |
ENSDART00000187744
|
rpl37
|
ribosomal protein L37 |
| chr21_+_20391978 | 1.00 |
ENSDART00000180817
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr25_-_24247584 | 1.00 |
ENSDART00000046349
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr22_+_2247645 | 1.00 |
ENSDART00000143366
ENSDART00000147852 |
si:dkey-4c15.8
|
si:dkey-4c15.8 |
| chr18_+_7553950 | 1.00 |
ENSDART00000193420
ENSDART00000062143 |
zgc:77650
|
zgc:77650 |
| chr10_+_158590 | 0.99 |
ENSDART00000081982
|
KCNJ15
|
potassium voltage-gated channel subfamily J member 15 |
| chr22_+_18929412 | 0.97 |
ENSDART00000161598
ENSDART00000166650 ENSDART00000015951 ENSDART00000105392 ENSDART00000131131 |
bsg
|
basigin |
| chr12_+_8569685 | 0.95 |
ENSDART00000031676
|
nrbf2b
|
nuclear receptor binding factor 2b |
| chr15_-_3534705 | 0.93 |
ENSDART00000158150
|
cog6
|
component of oligomeric golgi complex 6 |
| chr22_+_2751887 | 0.90 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr5_-_19014589 | 0.90 |
ENSDART00000002624
|
ranbp1
|
RAN binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CABZ01090890.1+atf6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 2.1 | 6.2 | GO:0034035 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 1.8 | 5.3 | GO:0046048 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 1.5 | 6.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 1.3 | 7.5 | GO:0072319 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 1.1 | 6.9 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 1.1 | 5.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.8 | 8.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.8 | 2.5 | GO:0032263 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.8 | 2.4 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.8 | 2.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.7 | 5.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.6 | 1.8 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.6 | 4.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.6 | 4.2 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.6 | 5.9 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.6 | 3.5 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.6 | 2.2 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.5 | 1.6 | GO:0019264 | L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 0.5 | 2.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.5 | 2.5 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.5 | 1.5 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.4 | 1.6 | GO:0060437 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.4 | 1.5 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.3 | 2.3 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.3 | 1.3 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.3 | 2.8 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.2 | 2.1 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.2 | 3.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 0.8 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.2 | 19.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.2 | 0.9 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.2 | 0.7 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.2 | 6.6 | GO:0030149 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.2 | 5.2 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.2 | 2.7 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.2 | 3.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.2 | 0.8 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 0.3 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 0.7 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.1 | 2.5 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 1.2 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.1 | 2.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.7 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.1 | 3.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 3.4 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 1.8 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 2.8 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 3.0 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.1 | 2.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.6 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 1.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 3.6 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.1 | 2.0 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.1 | 2.7 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.1 | 1.5 | GO:1990399 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.1 | 2.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 1.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 3.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 1.9 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 2.4 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 0.3 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 | 2.7 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.1 | 0.4 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.2 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 3.6 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.1 | 1.0 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 3.2 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.1 | 3.4 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.1 | 1.9 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.1 | 5.5 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 1.5 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 1.6 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 0.2 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 2.2 | GO:0051057 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 2.4 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 1.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 3.0 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.7 | GO:0003181 | atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 1.0 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.4 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 4.3 | GO:0043068 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 3.4 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.0 | 5.0 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 2.3 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.2 | GO:0050927 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.0 | 3.0 | GO:0046496 | pyridine nucleotide metabolic process(GO:0019362) nicotinamide nucleotide metabolic process(GO:0046496) |
| 0.0 | 0.7 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.5 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 3.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.6 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 3.8 | GO:0030900 | forebrain development(GO:0030900) |
| 0.0 | 0.7 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 1.4 | GO:0000723 | telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.0 | 2.2 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 1.9 | GO:0016125 | sterol metabolic process(GO:0016125) |
| 0.0 | 4.9 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 7.0 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 4.0 | GO:0006163 | purine nucleotide metabolic process(GO:0006163) |
| 0.0 | 0.4 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) muscle tissue morphogenesis(GO:0060415) |
| 0.0 | 3.2 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 1.5 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 1.7 | GO:0007163 | establishment or maintenance of cell polarity(GO:0007163) |
| 0.0 | 1.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.8 | GO:0006400 | tRNA modification(GO:0006400) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.3 | GO:0000938 | GARP complex(GO:0000938) |
| 0.5 | 4.7 | GO:0089701 | U2AF(GO:0089701) |
| 0.5 | 9.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.5 | 2.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.4 | 2.8 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.3 | 3.8 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.3 | 1.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.3 | 2.0 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 1.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 3.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 12.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.2 | 1.6 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 2.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 0.8 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 2.5 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.2 | 2.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 1.6 | GO:0030663 | COPI vesicle coat(GO:0030126) COPI-coated vesicle(GO:0030137) COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 1.2 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 1.0 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 1.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 4.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.7 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 4.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 6.2 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 4.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 15.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 6.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 32.7 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 1.4 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 2.7 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.5 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.0 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 3.7 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 4.4 | GO:0043235 | receptor complex(GO:0043235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 1.8 | 5.3 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 1.7 | 5.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 1.7 | 6.9 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 1.7 | 6.6 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 1.5 | 6.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 1.4 | 4.2 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 1.3 | 5.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 1.0 | 3.0 | GO:0052855 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 1.0 | 3.0 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.9 | 3.5 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.8 | 2.5 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.8 | 4.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.8 | 2.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.7 | 8.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.6 | 1.8 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.6 | 4.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.5 | 1.6 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.5 | 5.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.5 | 4.7 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.5 | 3.6 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.4 | 1.5 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.4 | 1.5 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.4 | 1.1 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.4 | 6.0 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.3 | 2.2 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.3 | 2.8 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.3 | 1.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.3 | 0.8 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.3 | 1.9 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 1.5 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 3.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 0.7 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.2 | 0.2 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.2 | 1.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 2.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.8 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.6 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.7 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 5.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 1.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 2.6 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 1.9 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 2.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.7 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 1.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 2.0 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.5 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 7.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.4 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 1.4 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 1.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 7.3 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 1.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 3.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 3.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 13.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.4 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 8.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 10.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.0 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 3.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 5.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.8 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 2.4 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.4 | GO:0004622 | lysophospholipase activity(GO:0004622) lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 1.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 2.1 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.4 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 2.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 6.6 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 1.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 3.4 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 3.9 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 8.5 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 4.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 4.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 1.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 2.7 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 2.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 2.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 2.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 3.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 3.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 12.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 11.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.2 | 2.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 1.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 4.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.2 | 5.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 5.4 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 3.8 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.1 | 1.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 4.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 3.0 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 1.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 2.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.0 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 1.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 2.7 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 4.9 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.9 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |