Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
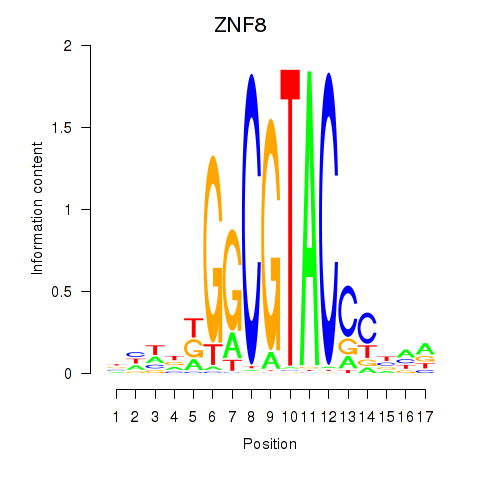
Results for ZNF8
Z-value: 0.38
Transcription factors associated with ZNF8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF8
|
ENSG00000083842.8 | zinc finger protein 8 |
|
ZNF8
|
ENSG00000273439.1 | zinc finger protein 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF8 | hg19_v2_chr19_+_58790314_58790358 | -0.70 | 1.2e-01 | Click! |
Activity profile of ZNF8 motif
Sorted Z-values of ZNF8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_104235229 | 0.16 |
ENST00000551650.1
|
RP11-650K20.3
|
Uncharacterized protein |
| chr11_+_46402482 | 0.13 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr8_-_61429315 | 0.10 |
ENST00000530725.1
ENST00000532232.1 |
RP11-163N6.2
|
RP11-163N6.2 |
| chr5_+_134210035 | 0.09 |
ENST00000506916.1
|
TXNDC15
|
thioredoxin domain containing 15 |
| chr6_-_26056695 | 0.08 |
ENST00000343677.2
|
HIST1H1C
|
histone cluster 1, H1c |
| chr6_+_29691056 | 0.07 |
ENST00000414333.1
ENST00000334668.4 ENST00000259951.7 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr1_-_186344802 | 0.07 |
ENST00000451586.1
|
TPR
|
translocated promoter region, nuclear basket protein |
| chr6_+_29691198 | 0.07 |
ENST00000440587.2
ENST00000434407.2 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr7_+_100303676 | 0.07 |
ENST00000303151.4
|
POP7
|
processing of precursor 7, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr6_+_31939608 | 0.06 |
ENST00000375331.2
ENST00000375333.2 |
STK19
|
serine/threonine kinase 19 |
| chr22_+_37415728 | 0.06 |
ENST00000404802.3
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr14_+_105941118 | 0.06 |
ENST00000550577.1
ENST00000538259.2 |
CRIP2
|
cysteine-rich protein 2 |
| chr1_-_55266926 | 0.06 |
ENST00000371276.4
|
TTC22
|
tetratricopeptide repeat domain 22 |
| chr5_-_43515125 | 0.06 |
ENST00000509489.1
|
C5orf34
|
chromosome 5 open reading frame 34 |
| chr22_+_37415676 | 0.06 |
ENST00000401419.3
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr22_+_37415700 | 0.06 |
ENST00000397129.1
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr19_+_35739782 | 0.06 |
ENST00000347609.4
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr22_-_37415475 | 0.06 |
ENST00000403892.3
ENST00000249042.3 ENST00000438203.1 |
TST
|
thiosulfate sulfurtransferase (rhodanese) |
| chr4_+_24797085 | 0.06 |
ENST00000382120.3
|
SOD3
|
superoxide dismutase 3, extracellular |
| chr8_+_38244638 | 0.06 |
ENST00000526356.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chrX_+_102631248 | 0.05 |
ENST00000361298.4
ENST00000372645.3 ENST00000372635.1 |
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr22_+_37415776 | 0.05 |
ENST00000341116.3
ENST00000429360.2 ENST00000404393.1 |
MPST
|
mercaptopyruvate sulfurtransferase |
| chr13_-_21750659 | 0.05 |
ENST00000400018.3
ENST00000314759.5 |
SKA3
|
spindle and kinetochore associated complex subunit 3 |
| chr19_-_5903714 | 0.05 |
ENST00000586349.1
ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12
NDUFA11
FUT5
|
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr1_-_246670614 | 0.04 |
ENST00000403792.3
|
SMYD3
|
SET and MYND domain containing 3 |
| chr13_-_25861530 | 0.04 |
ENST00000540661.1
|
MTMR6
|
myotubularin related protein 6 |
| chr7_+_1748798 | 0.04 |
ENST00000561626.1
|
ELFN1
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1 |
| chr16_-_53537105 | 0.04 |
ENST00000568596.1
ENST00000570004.1 ENST00000564497.1 ENST00000300245.4 ENST00000394657.7 |
AKTIP
|
AKT interacting protein |
| chr2_-_240322685 | 0.04 |
ENST00000544989.1
|
HDAC4
|
histone deacetylase 4 |
| chr19_+_35739897 | 0.04 |
ENST00000605618.1
ENST00000427250.1 ENST00000601623.1 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr4_-_77069573 | 0.04 |
ENST00000264883.3
|
NUP54
|
nucleoporin 54kDa |
| chr6_+_18155560 | 0.04 |
ENST00000546309.2
ENST00000388870.2 ENST00000397244.1 |
KDM1B
|
lysine (K)-specific demethylase 1B |
| chr2_+_169926047 | 0.03 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr1_-_235292250 | 0.03 |
ENST00000366607.4
|
TOMM20
|
translocase of outer mitochondrial membrane 20 homolog (yeast) |
| chr7_+_117824086 | 0.03 |
ENST00000249299.2
ENST00000424702.1 |
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr4_-_186347099 | 0.03 |
ENST00000505357.1
ENST00000264689.6 |
UFSP2
|
UFM1-specific peptidase 2 |
| chr12_-_74686314 | 0.03 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr17_+_48624450 | 0.03 |
ENST00000006658.6
ENST00000356488.4 ENST00000393244.3 |
SPATA20
|
spermatogenesis associated 20 |
| chr10_-_73611046 | 0.03 |
ENST00000394934.1
ENST00000394936.3 |
PSAP
|
prosaposin |
| chr17_+_7517264 | 0.03 |
ENST00000593717.1
ENST00000572182.1 ENST00000574539.1 ENST00000576728.1 ENST00000575314.1 ENST00000570547.1 ENST00000572262.1 ENST00000576478.1 |
AC007421.1
SHBG
|
AC007421.1 sex hormone-binding globulin |
| chr11_+_46402583 | 0.03 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr9_-_37576226 | 0.02 |
ENST00000432825.2
|
FBXO10
|
F-box protein 10 |
| chr2_-_172290482 | 0.02 |
ENST00000442541.1
ENST00000392599.2 ENST00000375258.4 |
METTL8
|
methyltransferase like 8 |
| chr6_-_116601044 | 0.02 |
ENST00000368608.3
|
TSPYL1
|
TSPY-like 1 |
| chr9_+_1050331 | 0.02 |
ENST00000382255.3
ENST00000382251.3 ENST00000412350.2 |
DMRT2
|
doublesex and mab-3 related transcription factor 2 |
| chr5_-_93447333 | 0.02 |
ENST00000395965.3
ENST00000505869.1 ENST00000509163.1 |
FAM172A
|
family with sequence similarity 172, member A |
| chr8_+_61429416 | 0.02 |
ENST00000262646.7
ENST00000531289.1 |
RAB2A
|
RAB2A, member RAS oncogene family |
| chr11_-_58345569 | 0.02 |
ENST00000528954.1
ENST00000528489.1 |
LPXN
|
leupaxin |
| chr11_+_69455855 | 0.02 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr16_+_29973351 | 0.02 |
ENST00000602948.1
ENST00000279396.6 ENST00000575829.2 ENST00000561899.2 |
TMEM219
|
transmembrane protein 219 |
| chr2_-_105946491 | 0.02 |
ENST00000393359.2
|
TGFBRAP1
|
transforming growth factor, beta receptor associated protein 1 |
| chr22_+_30163340 | 0.02 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr1_+_25664408 | 0.02 |
ENST00000374358.4
|
TMEM50A
|
transmembrane protein 50A |
| chr2_+_172290707 | 0.02 |
ENST00000375255.3
ENST00000539783.1 |
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr16_+_21964662 | 0.02 |
ENST00000561553.1
ENST00000565331.1 |
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II |
| chr1_+_186344883 | 0.01 |
ENST00000367470.3
|
C1orf27
|
chromosome 1 open reading frame 27 |
| chr5_+_102455968 | 0.01 |
ENST00000358359.3
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr13_+_53029564 | 0.01 |
ENST00000468284.1
ENST00000378034.3 ENST00000258607.5 ENST00000378037.5 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr19_+_52074502 | 0.01 |
ENST00000545217.1
ENST00000262259.2 ENST00000596504.1 |
ZNF175
|
zinc finger protein 175 |
| chr11_-_46638720 | 0.01 |
ENST00000326737.3
|
HARBI1
|
harbinger transposase derived 1 |
| chr6_+_88117683 | 0.01 |
ENST00000369562.4
|
C6ORF165
|
UPF0704 protein C6orf165 |
| chr11_-_46638378 | 0.01 |
ENST00000529192.1
|
HARBI1
|
harbinger transposase derived 1 |
| chr6_+_53659746 | 0.01 |
ENST00000370888.1
|
LRRC1
|
leucine rich repeat containing 1 |
| chr1_-_100715372 | 0.01 |
ENST00000370131.3
ENST00000370132.4 |
DBT
|
dihydrolipoamide branched chain transacylase E2 |
| chr14_+_51706886 | 0.01 |
ENST00000457354.2
|
TMX1
|
thioredoxin-related transmembrane protein 1 |
| chr6_+_106959718 | 0.01 |
ENST00000369066.3
|
AIM1
|
absent in melanoma 1 |
| chr17_+_48585902 | 0.01 |
ENST00000452039.1
|
MYCBPAP
|
MYCBP associated protein |
| chr19_-_55652290 | 0.01 |
ENST00000589745.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr1_+_211500129 | 0.01 |
ENST00000427925.2
ENST00000261464.5 |
TRAF5
|
TNF receptor-associated factor 5 |
| chr1_-_246670519 | 0.01 |
ENST00000388985.4
ENST00000490107.1 |
SMYD3
|
SET and MYND domain containing 3 |
| chr8_-_109260897 | 0.01 |
ENST00000521297.1
ENST00000519030.1 ENST00000521440.1 ENST00000518345.1 ENST00000519627.1 ENST00000220849.5 |
EIF3E
|
eukaryotic translation initiation factor 3, subunit E |
| chr1_+_186344945 | 0.01 |
ENST00000419367.3
ENST00000287859.6 |
C1orf27
|
chromosome 1 open reading frame 27 |
| chr1_+_6615241 | 0.01 |
ENST00000333172.6
ENST00000328191.4 ENST00000351136.3 |
TAS1R1
|
taste receptor, type 1, member 1 |
| chrX_+_1387693 | 0.01 |
ENST00000381529.3
ENST00000432318.2 ENST00000361536.3 ENST00000501036.2 ENST00000381524.3 ENST00000412290.1 |
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr1_-_228296956 | 0.01 |
ENST00000366744.1
ENST00000348259.5 ENST00000366747.3 ENST00000366746.3 ENST00000295008.4 ENST00000336520.3 ENST00000366731.5 ENST00000411464.2 ENST00000457264.1 ENST00000336300.5 ENST00000430433.1 ENST00000391867.3 |
MRPL55
|
mitochondrial ribosomal protein L55 |
| chr16_+_68573116 | 0.00 |
ENST00000570495.1
ENST00000563169.2 ENST00000564323.1 ENST00000562156.1 ENST00000573685.1 |
ZFP90
|
ZFP90 zinc finger protein |
| chr12_+_52463751 | 0.00 |
ENST00000336854.4
ENST00000550604.1 ENST00000553049.1 ENST00000548915.1 |
C12orf44
|
chromosome 12 open reading frame 44 |
| chr2_+_87144738 | 0.00 |
ENST00000559485.1
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr9_-_32552551 | 0.00 |
ENST00000360538.2
ENST00000379858.1 |
TOPORS
|
topoisomerase I binding, arginine/serine-rich, E3 ubiquitin protein ligase |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0009440 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.0 | 0.2 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0045799 | negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 0.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |