Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
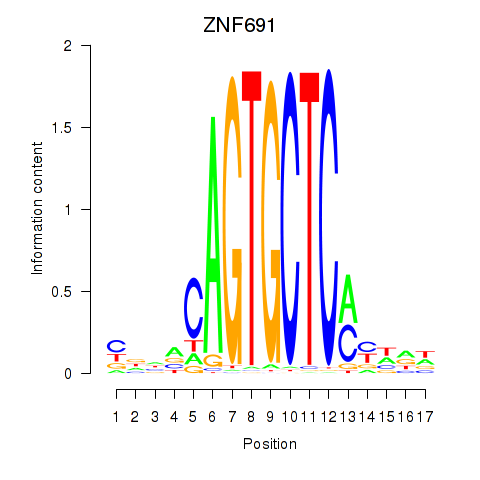
Results for ZNF691
Z-value: 0.66
Transcription factors associated with ZNF691
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF691
|
ENSG00000164011.13 | zinc finger protein 691 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF691 | hg19_v2_chr1_+_43312258_43312310 | -0.48 | 3.4e-01 | Click! |
Activity profile of ZNF691 motif
Sorted Z-values of ZNF691 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_40516560 | 0.44 |
ENST00000513473.1
|
RBM47
|
RNA binding motif protein 47 |
| chr3_+_177534653 | 0.39 |
ENST00000436078.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr19_+_40877583 | 0.38 |
ENST00000596470.1
|
PLD3
|
phospholipase D family, member 3 |
| chr10_+_4828815 | 0.38 |
ENST00000533295.1
|
AKR1E2
|
aldo-keto reductase family 1, member E2 |
| chr22_+_23264766 | 0.32 |
ENST00000390331.2
|
IGLC7
|
immunoglobulin lambda constant 7 |
| chr11_+_102188272 | 0.30 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr20_-_1317555 | 0.28 |
ENST00000537552.1
|
AL136531.1
|
HCG2043693; Uncharacterized protein |
| chr22_-_32058416 | 0.28 |
ENST00000439502.2
|
PISD
|
phosphatidylserine decarboxylase |
| chr19_-_5903714 | 0.28 |
ENST00000586349.1
ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12
NDUFA11
FUT5
|
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr15_-_74726283 | 0.27 |
ENST00000543145.2
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr15_-_40398812 | 0.27 |
ENST00000561360.1
|
BMF
|
Bcl2 modifying factor |
| chr15_-_86338100 | 0.27 |
ENST00000536947.1
|
KLHL25
|
kelch-like family member 25 |
| chr8_-_145159083 | 0.26 |
ENST00000398712.2
|
SHARPIN
|
SHANK-associated RH domain interactor |
| chr8_-_144679264 | 0.26 |
ENST00000531953.1
ENST00000526133.1 |
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr3_-_107596910 | 0.25 |
ENST00000464359.2
ENST00000464823.1 ENST00000466155.1 ENST00000473528.2 ENST00000608306.1 ENST00000488852.1 ENST00000608137.1 ENST00000608307.1 ENST00000609429.1 ENST00000601385.1 ENST00000475362.1 ENST00000600240.1 ENST00000600749.1 |
LINC00635
|
long intergenic non-protein coding RNA 635 |
| chr19_+_40873617 | 0.24 |
ENST00000599353.1
|
PLD3
|
phospholipase D family, member 3 |
| chr9_-_135754164 | 0.24 |
ENST00000298545.3
|
AK8
|
adenylate kinase 8 |
| chr2_-_203736452 | 0.22 |
ENST00000419460.1
|
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr17_+_38121772 | 0.21 |
ENST00000577447.1
|
GSDMA
|
gasdermin A |
| chr17_-_61777459 | 0.21 |
ENST00000578993.1
ENST00000583211.1 ENST00000259006.3 |
LIMD2
|
LIM domain containing 2 |
| chr12_+_120502441 | 0.21 |
ENST00000446727.2
|
CCDC64
|
coiled-coil domain containing 64 |
| chr16_+_810728 | 0.19 |
ENST00000563941.1
ENST00000545450.2 ENST00000566549.1 |
MSLN
|
mesothelin |
| chr5_+_53686658 | 0.19 |
ENST00000512618.1
|
LINC01033
|
long intergenic non-protein coding RNA 1033 |
| chr14_+_55033815 | 0.19 |
ENST00000554335.1
|
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr8_+_27169138 | 0.19 |
ENST00000522338.1
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr21_-_37852359 | 0.18 |
ENST00000399137.1
ENST00000399135.1 |
CLDN14
|
claudin 14 |
| chr8_-_102181718 | 0.18 |
ENST00000565617.1
|
KB-1460A1.5
|
KB-1460A1.5 |
| chr9_+_135754263 | 0.17 |
ENST00000356311.5
ENST00000350499.6 |
C9orf9
|
chromosome 9 open reading frame 9 |
| chr21_+_44866471 | 0.16 |
ENST00000448049.1
|
LINC00319
|
long intergenic non-protein coding RNA 319 |
| chr1_+_26856236 | 0.16 |
ENST00000374168.2
ENST00000374166.4 |
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr5_+_3596168 | 0.15 |
ENST00000302006.3
|
IRX1
|
iroquois homeobox 1 |
| chr20_-_2644832 | 0.14 |
ENST00000380851.5
ENST00000380843.4 |
IDH3B
|
isocitrate dehydrogenase 3 (NAD+) beta |
| chr6_+_31021225 | 0.14 |
ENST00000565192.1
ENST00000562344.1 |
HCG22
|
HLA complex group 22 |
| chrX_-_153775760 | 0.14 |
ENST00000440967.1
ENST00000393564.2 ENST00000369620.2 |
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr15_-_39486510 | 0.14 |
ENST00000560743.1
|
RP11-265N7.1
|
RP11-265N7.1 |
| chr5_-_179107975 | 0.13 |
ENST00000376974.4
|
CBY3
|
chibby homolog 3 (Drosophila) |
| chr10_+_104678032 | 0.13 |
ENST00000369878.4
ENST00000369875.3 |
CNNM2
|
cyclin M2 |
| chr8_-_123793048 | 0.13 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr2_+_202098203 | 0.13 |
ENST00000450491.1
ENST00000440732.1 ENST00000392258.3 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr5_-_176889381 | 0.13 |
ENST00000393563.4
ENST00000512501.1 |
DBN1
|
drebrin 1 |
| chr1_-_33647267 | 0.13 |
ENST00000291416.5
|
TRIM62
|
tripartite motif containing 62 |
| chr19_-_59023348 | 0.13 |
ENST00000601355.1
ENST00000263093.2 |
SLC27A5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr2_-_74669009 | 0.12 |
ENST00000272430.5
|
RTKN
|
rhotekin |
| chr17_-_80797886 | 0.12 |
ENST00000572562.1
|
ZNF750
|
zinc finger protein 750 |
| chr3_+_9850199 | 0.12 |
ENST00000452597.1
|
TTLL3
|
tubulin tyrosine ligase-like family, member 3 |
| chr14_+_55034330 | 0.12 |
ENST00000251091.5
|
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr22_-_32058166 | 0.12 |
ENST00000435900.1
ENST00000336566.4 |
PISD
|
phosphatidylserine decarboxylase |
| chr17_-_73840614 | 0.11 |
ENST00000586108.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr8_+_32579321 | 0.11 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr15_+_84908573 | 0.11 |
ENST00000424966.1
ENST00000422563.2 |
GOLGA6L4
|
golgin A6 family-like 4 |
| chrX_+_153672468 | 0.11 |
ENST00000393600.3
|
FAM50A
|
family with sequence similarity 50, member A |
| chr11_-_65655906 | 0.11 |
ENST00000533045.1
ENST00000338369.2 ENST00000357519.4 |
FIBP
|
fibroblast growth factor (acidic) intracellular binding protein |
| chrX_+_153775821 | 0.10 |
ENST00000263518.6
ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr4_-_120243545 | 0.10 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr2_-_203736334 | 0.10 |
ENST00000392237.2
ENST00000416760.1 ENST00000412210.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr17_+_80416482 | 0.10 |
ENST00000309794.11
ENST00000345415.7 ENST00000457415.3 ENST00000584411.1 ENST00000412079.2 ENST00000577432.1 |
NARF
|
nuclear prelamin A recognition factor |
| chr15_-_49912944 | 0.10 |
ENST00000559905.1
|
FAM227B
|
family with sequence similarity 227, member B |
| chr8_-_119964434 | 0.10 |
ENST00000297350.4
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b |
| chr8_+_27168988 | 0.10 |
ENST00000397501.1
ENST00000338238.4 ENST00000544172.1 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr19_-_893200 | 0.09 |
ENST00000269814.4
ENST00000395808.3 ENST00000312090.6 ENST00000325464.1 |
MED16
|
mediator complex subunit 16 |
| chr7_-_84569561 | 0.09 |
ENST00000439105.1
|
AC074183.4
|
AC074183.4 |
| chr17_+_36908984 | 0.09 |
ENST00000225426.4
ENST00000579088.1 |
PSMB3
|
proteasome (prosome, macropain) subunit, beta type, 3 |
| chr14_+_103388976 | 0.09 |
ENST00000299155.5
|
AMN
|
amnion associated transmembrane protein |
| chr8_-_144679296 | 0.09 |
ENST00000317198.6
|
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr19_-_15560730 | 0.09 |
ENST00000389282.4
ENST00000263381.7 |
WIZ
|
widely interspaced zinc finger motifs |
| chr1_+_145516560 | 0.08 |
ENST00000537888.1
|
PEX11B
|
peroxisomal biogenesis factor 11 beta |
| chr2_+_68962014 | 0.08 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr9_+_139553306 | 0.08 |
ENST00000371699.1
|
EGFL7
|
EGF-like-domain, multiple 7 |
| chr16_+_53738053 | 0.08 |
ENST00000394647.3
|
FTO
|
fat mass and obesity associated |
| chr8_-_144679532 | 0.08 |
ENST00000534380.1
ENST00000533494.1 ENST00000531218.1 ENST00000526340.1 ENST00000533204.1 ENST00000532400.1 ENST00000529516.1 ENST00000534377.1 ENST00000531621.1 ENST00000530191.1 ENST00000524900.1 ENST00000526838.1 ENST00000531931.1 ENST00000534475.1 ENST00000442189.2 ENST00000524624.1 ENST00000532596.1 ENST00000529832.1 ENST00000530306.1 ENST00000530545.1 ENST00000525261.1 ENST00000534804.1 ENST00000528303.1 ENST00000528610.1 |
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr11_+_64949899 | 0.08 |
ENST00000531068.1
ENST00000527699.1 ENST00000533909.1 ENST00000527323.1 |
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr15_-_100258029 | 0.08 |
ENST00000378904.2
|
DKFZP779J2370
|
DKFZP779J2370 |
| chr6_-_144329531 | 0.08 |
ENST00000429150.1
ENST00000392309.1 ENST00000416623.1 ENST00000392307.1 |
PLAGL1
|
pleiomorphic adenoma gene-like 1 |
| chr18_+_12947126 | 0.07 |
ENST00000592170.1
|
SEH1L
|
SEH1-like (S. cerevisiae) |
| chr22_-_43539346 | 0.07 |
ENST00000327555.5
ENST00000290429.6 |
MCAT
|
malonyl CoA:ACP acyltransferase (mitochondrial) |
| chr12_+_27619743 | 0.07 |
ENST00000298876.4
ENST00000416383.1 |
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr8_+_145294015 | 0.07 |
ENST00000544576.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr16_-_1821496 | 0.07 |
ENST00000564628.1
ENST00000563498.1 |
NME3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr9_+_139847347 | 0.07 |
ENST00000371632.3
|
LCN12
|
lipocalin 12 |
| chr20_-_33735070 | 0.07 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chrX_-_153775047 | 0.06 |
ENST00000433845.1
ENST00000439227.1 |
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr1_+_11994715 | 0.06 |
ENST00000449038.1
ENST00000376369.3 ENST00000429000.2 ENST00000196061.4 |
PLOD1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr8_+_32579341 | 0.06 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr20_-_45976816 | 0.06 |
ENST00000441977.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr1_+_28199047 | 0.06 |
ENST00000373925.1
ENST00000328928.7 ENST00000373927.3 ENST00000427466.1 ENST00000442118.1 ENST00000373921.3 |
THEMIS2
|
thymocyte selection associated family member 2 |
| chr5_+_7396141 | 0.06 |
ENST00000338316.4
|
ADCY2
|
adenylate cyclase 2 (brain) |
| chr3_-_194968576 | 0.06 |
ENST00000429994.1
|
XXYLT1
|
xyloside xylosyltransferase 1 |
| chr17_+_80416609 | 0.05 |
ENST00000577410.1
|
NARF
|
nuclear prelamin A recognition factor |
| chr15_-_26108355 | 0.05 |
ENST00000356865.6
|
ATP10A
|
ATPase, class V, type 10A |
| chr17_+_37026106 | 0.05 |
ENST00000318008.6
|
LASP1
|
LIM and SH3 protein 1 |
| chr4_+_657485 | 0.05 |
ENST00000471824.2
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr1_-_233431458 | 0.05 |
ENST00000258229.9
ENST00000430153.1 |
PCNXL2
|
pecanex-like 2 (Drosophila) |
| chr10_-_99258135 | 0.05 |
ENST00000327238.10
ENST00000327277.7 ENST00000355839.6 ENST00000437002.1 ENST00000422685.1 |
MMS19
|
MMS19 nucleotide excision repair homolog (S. cerevisiae) |
| chr17_-_3417062 | 0.05 |
ENST00000570318.1
ENST00000541913.1 |
SPATA22
|
spermatogenesis associated 22 |
| chr2_+_68961934 | 0.05 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr3_-_157251383 | 0.05 |
ENST00000487753.1
ENST00000489602.1 ENST00000461299.1 ENST00000479987.1 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr11_-_133826852 | 0.04 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chr1_-_148202536 | 0.04 |
ENST00000544708.1
|
PPIAL4D
|
peptidylprolyl isomerase A (cyclophilin A)-like 4D |
| chr17_-_41738931 | 0.04 |
ENST00000329168.3
ENST00000549132.1 |
MEOX1
|
mesenchyme homeobox 1 |
| chr6_-_90529418 | 0.04 |
ENST00000439638.1
ENST00000369393.3 ENST00000428876.1 |
MDN1
|
MDN1, midasin homolog (yeast) |
| chr6_+_31553901 | 0.04 |
ENST00000418507.2
ENST00000438075.2 ENST00000376100.3 ENST00000376111.4 |
LST1
|
leukocyte specific transcript 1 |
| chr12_-_10605929 | 0.04 |
ENST00000347831.5
ENST00000359151.3 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr6_+_151186554 | 0.04 |
ENST00000367321.3
ENST00000367307.4 |
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr11_-_36619771 | 0.04 |
ENST00000311485.3
ENST00000527033.1 ENST00000532616.1 |
RAG2
|
recombination activating gene 2 |
| chr19_+_50084561 | 0.04 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr19_+_50380917 | 0.04 |
ENST00000535102.2
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr2_+_68961905 | 0.03 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr12_-_51717948 | 0.03 |
ENST00000267012.4
|
BIN2
|
bridging integrator 2 |
| chr17_-_73839792 | 0.03 |
ENST00000590762.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr12_-_109221160 | 0.03 |
ENST00000326470.5
|
SSH1
|
slingshot protein phosphatase 1 |
| chr17_+_80416050 | 0.03 |
ENST00000579198.1
ENST00000390006.4 ENST00000580296.1 |
NARF
|
nuclear prelamin A recognition factor |
| chr17_+_37026284 | 0.03 |
ENST00000433206.2
ENST00000435347.3 |
LASP1
|
LIM and SH3 protein 1 |
| chr12_-_51717922 | 0.03 |
ENST00000452142.2
|
BIN2
|
bridging integrator 2 |
| chr3_-_46037299 | 0.03 |
ENST00000296137.2
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr12_-_51717875 | 0.03 |
ENST00000604560.1
|
BIN2
|
bridging integrator 2 |
| chrX_-_62571187 | 0.03 |
ENST00000335144.3
|
SPIN4
|
spindlin family, member 4 |
| chr7_-_143059780 | 0.03 |
ENST00000409578.1
ENST00000409346.1 |
FAM131B
|
family with sequence similarity 131, member B |
| chr10_+_104678102 | 0.03 |
ENST00000433628.2
|
CNNM2
|
cyclin M2 |
| chr6_+_31553978 | 0.02 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr9_+_116267536 | 0.02 |
ENST00000374136.1
|
RGS3
|
regulator of G-protein signaling 3 |
| chr15_+_89402148 | 0.02 |
ENST00000560601.1
|
ACAN
|
aggrecan |
| chr20_+_62887081 | 0.02 |
ENST00000369758.4
ENST00000299468.7 ENST00000609372.1 ENST00000610196.1 ENST00000308824.6 |
PCMTD2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr19_-_17932314 | 0.02 |
ENST00000598577.1
ENST00000317306.7 ENST00000379695.5 |
INSL3
|
insulin-like 3 (Leydig cell) |
| chr2_+_106468204 | 0.02 |
ENST00000425756.1
ENST00000393349.2 |
NCK2
|
NCK adaptor protein 2 |
| chr12_-_51718436 | 0.02 |
ENST00000544402.1
|
BIN2
|
bridging integrator 2 |
| chr3_+_37493590 | 0.02 |
ENST00000422441.1
|
ITGA9
|
integrin, alpha 9 |
| chr10_-_4720301 | 0.02 |
ENST00000449712.1
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr17_+_75401152 | 0.02 |
ENST00000585930.1
|
SEPT9
|
septin 9 |
| chr6_-_27279949 | 0.02 |
ENST00000444565.1
ENST00000377451.2 |
POM121L2
|
POM121 transmembrane nucleoporin-like 2 |
| chr16_-_11492366 | 0.02 |
ENST00000595360.1
|
CTD-3088G3.8
|
Protein LOC388210 |
| chr17_-_73840415 | 0.02 |
ENST00000592386.1
ENST00000412096.2 ENST00000586147.1 |
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr1_+_178482212 | 0.02 |
ENST00000319416.2
ENST00000258298.2 ENST00000367643.3 ENST00000367642.3 |
TEX35
|
testis expressed 35 |
| chr7_+_7811992 | 0.02 |
ENST00000406829.1
|
RPA3-AS1
|
RPA3 antisense RNA 1 |
| chr1_+_39796810 | 0.02 |
ENST00000289893.4
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chrX_+_102469997 | 0.02 |
ENST00000372695.5
ENST00000372691.3 |
BEX4
|
brain expressed, X-linked 4 |
| chr11_-_64889529 | 0.01 |
ENST00000531743.1
ENST00000527548.1 ENST00000526555.1 ENST00000279259.3 |
FAU
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed |
| chr6_-_159240415 | 0.01 |
ENST00000367075.3
|
EZR
|
ezrin |
| chr14_-_74296806 | 0.01 |
ENST00000555539.1
|
RP5-1021I20.2
|
RP5-1021I20.2 |
| chr17_-_38821373 | 0.01 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr1_+_90308981 | 0.01 |
ENST00000527156.1
|
LRRC8D
|
leucine rich repeat containing 8 family, member D |
| chr3_+_33331156 | 0.01 |
ENST00000542085.1
|
FBXL2
|
F-box and leucine-rich repeat protein 2 |
| chr5_-_131347501 | 0.01 |
ENST00000543479.1
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr22_-_45608237 | 0.01 |
ENST00000492273.1
|
KIAA0930
|
KIAA0930 |
| chr1_-_155232221 | 0.00 |
ENST00000355379.3
|
SCAMP3
|
secretory carrier membrane protein 3 |
| chr19_-_9649303 | 0.00 |
ENST00000253115.2
|
ZNF426
|
zinc finger protein 426 |
| chr15_-_58571445 | 0.00 |
ENST00000558231.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr7_+_134576317 | 0.00 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr17_+_45286387 | 0.00 |
ENST00000572316.1
ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF691
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:2000537 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.2 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.3 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.3 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.3 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0052053 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.3 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.3 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.0 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.1 | 0.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.4 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.3 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.2 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.1 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.0 | 0.1 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | ST GA12 PATHWAY | G alpha 12 Pathway |