Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
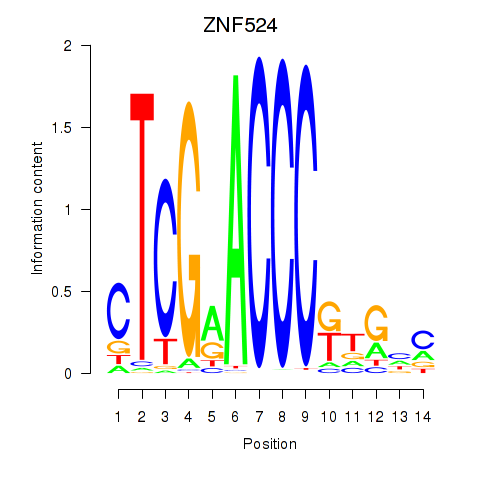
Results for ZNF524
Z-value: 1.08
Transcription factors associated with ZNF524
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF524
|
ENSG00000171443.6 | zinc finger protein 524 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF524 | hg19_v2_chr19_+_56111680_56111735 | 0.64 | 1.7e-01 | Click! |
Activity profile of ZNF524 motif
Sorted Z-values of ZNF524 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_54372877 | 3.89 |
ENST00000414489.1
|
MYADM
|
myeloid-associated differentiation marker |
| chr19_+_36134528 | 0.87 |
ENST00000591135.1
|
ETV2
|
ets variant 2 |
| chr19_+_50015870 | 0.85 |
ENST00000599701.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr7_-_155437075 | 0.83 |
ENST00000401694.1
|
AC009403.2
|
Protein LOC100506302 |
| chr14_+_105559784 | 0.75 |
ENST00000548104.1
|
RP11-44N21.1
|
RP11-44N21.1 |
| chr17_+_7253667 | 0.57 |
ENST00000570504.1
ENST00000574499.1 |
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr19_-_54984354 | 0.53 |
ENST00000301200.2
|
CDC42EP5
|
CDC42 effector protein (Rho GTPase binding) 5 |
| chr5_-_73936544 | 0.51 |
ENST00000509127.2
|
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr17_-_79304150 | 0.49 |
ENST00000574093.1
|
TMEM105
|
transmembrane protein 105 |
| chr19_+_50016411 | 0.44 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr2_+_113342011 | 0.43 |
ENST00000324913.5
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr19_-_12708113 | 0.42 |
ENST00000440366.1
|
ZNF490
|
zinc finger protein 490 |
| chr2_+_136499287 | 0.42 |
ENST00000415164.1
|
UBXN4
|
UBX domain protein 4 |
| chr19_-_11039261 | 0.41 |
ENST00000590329.1
ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2
|
Yip1 domain family, member 2 |
| chr8_+_145137489 | 0.40 |
ENST00000355091.4
ENST00000525087.1 ENST00000361036.6 ENST00000524418.1 |
GPAA1
|
glycosylphosphatidylinositol anchor attachment 1 |
| chr22_-_37415475 | 0.40 |
ENST00000403892.3
ENST00000249042.3 ENST00000438203.1 |
TST
|
thiosulfate sulfurtransferase (rhodanese) |
| chr16_-_75467274 | 0.39 |
ENST00000566254.1
|
CFDP1
|
craniofacial development protein 1 |
| chrY_+_15418467 | 0.38 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chrX_-_48937684 | 0.37 |
ENST00000465382.1
ENST00000423215.2 |
WDR45
|
WD repeat domain 45 |
| chr10_+_134000404 | 0.37 |
ENST00000338492.4
ENST00000368629.1 |
DPYSL4
|
dihydropyrimidinase-like 4 |
| chr19_-_11039188 | 0.36 |
ENST00000588347.1
|
YIPF2
|
Yip1 domain family, member 2 |
| chr19_-_36643329 | 0.36 |
ENST00000589154.1
|
COX7A1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr8_+_144798429 | 0.35 |
ENST00000338033.4
ENST00000395107.4 ENST00000395108.2 |
MAPK15
|
mitogen-activated protein kinase 15 |
| chr12_+_113354341 | 0.35 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr19_+_56159509 | 0.34 |
ENST00000586790.1
ENST00000591578.1 ENST00000588740.1 |
CCDC106
|
coiled-coil domain containing 106 |
| chr19_-_11266437 | 0.33 |
ENST00000586708.1
ENST00000591396.1 ENST00000592967.1 ENST00000585486.1 ENST00000585567.1 |
SPC24
|
SPC24, NDC80 kinetochore complex component |
| chr10_+_4828815 | 0.33 |
ENST00000533295.1
|
AKR1E2
|
aldo-keto reductase family 1, member E2 |
| chr16_-_25122735 | 0.33 |
ENST00000563176.1
|
RP11-449H11.1
|
RP11-449H11.1 |
| chr1_-_19229218 | 0.32 |
ENST00000432718.1
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr11_-_65314905 | 0.31 |
ENST00000527339.1
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr11_+_810221 | 0.31 |
ENST00000530398.1
|
RPLP2
|
ribosomal protein, large, P2 |
| chr2_-_20101385 | 0.31 |
ENST00000431392.1
|
TTC32
|
tetratricopeptide repeat domain 32 |
| chr22_-_46646576 | 0.30 |
ENST00000314567.3
|
CDPF1
|
cysteine-rich, DPF motif domain containing 1 |
| chr17_+_7239821 | 0.30 |
ENST00000158762.3
ENST00000570457.2 |
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr16_-_2205352 | 0.30 |
ENST00000563192.1
|
RP11-304L19.5
|
RP11-304L19.5 |
| chr19_+_1041187 | 0.30 |
ENST00000531467.1
|
ABCA7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr16_-_75284758 | 0.29 |
ENST00000561970.1
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr8_-_11324273 | 0.29 |
ENST00000284486.4
|
FAM167A
|
family with sequence similarity 167, member A |
| chr16_+_1543337 | 0.29 |
ENST00000262319.6
|
TELO2
|
telomere maintenance 2 |
| chr13_-_21348066 | 0.29 |
ENST00000382758.1
|
N6AMT2
|
N-6 adenine-specific DNA methyltransferase 2 (putative) |
| chr17_+_78965624 | 0.29 |
ENST00000325167.5
|
CHMP6
|
charged multivesicular body protein 6 |
| chr17_-_43339474 | 0.29 |
ENST00000331780.4
|
SPATA32
|
spermatogenesis associated 32 |
| chr17_-_80170689 | 0.27 |
ENST00000389641.4
ENST00000392347.1 ENST00000392343.3 |
CCDC57
|
coiled-coil domain containing 57 |
| chr19_-_44174305 | 0.27 |
ENST00000601723.1
ENST00000339082.3 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chr2_+_71205510 | 0.26 |
ENST00000272421.6
ENST00000441349.1 ENST00000457410.1 |
ANKRD53
|
ankyrin repeat domain 53 |
| chr17_-_79533608 | 0.26 |
ENST00000572760.1
ENST00000573876.1 |
NPLOC4
|
nuclear protein localization 4 homolog (S. cerevisiae) |
| chr2_-_24149918 | 0.26 |
ENST00000439915.1
|
ATAD2B
|
ATPase family, AAA domain containing 2B |
| chr19_+_13228917 | 0.26 |
ENST00000586171.1
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr17_+_48351785 | 0.25 |
ENST00000507382.1
|
TMEM92
|
transmembrane protein 92 |
| chr19_-_460996 | 0.25 |
ENST00000264554.6
|
SHC2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr12_+_12202785 | 0.25 |
ENST00000586576.1
ENST00000464885.2 |
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr19_+_50016610 | 0.25 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr16_-_12184159 | 0.25 |
ENST00000312019.2
|
RP11-276H1.3
|
RP11-276H1.3 |
| chrY_-_297445 | 0.25 |
ENSTR0000390665.3
|
PPP2R3B
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr4_-_74964904 | 0.25 |
ENST00000508487.2
|
CXCL2
|
chemokine (C-X-C motif) ligand 2 |
| chr19_-_11266471 | 0.25 |
ENST00000592540.1
|
SPC24
|
SPC24, NDC80 kinetochore complex component |
| chr19_+_35899569 | 0.25 |
ENST00000600405.1
|
AC002511.1
|
AC002511.1 |
| chr3_+_32433363 | 0.25 |
ENST00000465248.1
|
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr15_+_64680003 | 0.24 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr16_+_812506 | 0.24 |
ENST00000569566.1
|
MSLN
|
mesothelin |
| chr15_+_96897466 | 0.24 |
ENST00000558382.1
ENST00000558499.1 |
RP11-522B15.3
|
RP11-522B15.3 |
| chr14_+_77425972 | 0.24 |
ENST00000553613.1
|
RP11-7F17.7
|
RP11-7F17.7 |
| chr22_+_37415728 | 0.24 |
ENST00000404802.3
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr11_+_65686952 | 0.23 |
ENST00000527119.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr10_-_102289611 | 0.23 |
ENST00000299166.4
ENST00000370320.4 ENST00000531258.1 ENST00000370322.1 ENST00000535773.1 |
NDUFB8
SEC31B
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8, 19kDa SEC31 homolog B (S. cerevisiae) |
| chr19_+_49496705 | 0.22 |
ENST00000595090.1
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr7_+_100271446 | 0.22 |
ENST00000419828.1
ENST00000427895.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr22_+_42470289 | 0.22 |
ENST00000419475.1
|
FAM109B
|
family with sequence similarity 109, member B |
| chr19_-_44174330 | 0.22 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr17_-_8079648 | 0.22 |
ENST00000449985.2
ENST00000532998.1 ENST00000437139.2 ENST00000533070.1 ENST00000316425.5 |
TMEM107
|
transmembrane protein 107 |
| chr16_+_2097970 | 0.22 |
ENST00000382538.6
ENST00000401874.2 ENST00000353929.4 |
TSC2
|
tuberous sclerosis 2 |
| chr1_+_171060018 | 0.22 |
ENST00000367755.4
ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3
|
flavin containing monooxygenase 3 |
| chr6_+_139349903 | 0.22 |
ENST00000461027.1
|
ABRACL
|
ABRA C-terminal like |
| chr7_+_100209725 | 0.22 |
ENST00000223054.4
|
MOSPD3
|
motile sperm domain containing 3 |
| chr2_-_152118276 | 0.22 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr15_+_67835517 | 0.22 |
ENST00000395476.2
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr7_+_99156393 | 0.22 |
ENST00000422164.1
ENST00000422647.1 ENST00000427931.1 |
ZNF655
|
zinc finger protein 655 |
| chr1_-_161147275 | 0.22 |
ENST00000319769.5
ENST00000367998.1 |
B4GALT3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr19_-_12624647 | 0.22 |
ENST00000455490.1
|
ZNF709
|
zinc finger protein 709 |
| chr17_+_42422629 | 0.22 |
ENST00000589536.1
ENST00000587109.1 ENST00000587518.1 |
GRN
|
granulin |
| chr1_+_3370990 | 0.21 |
ENST00000378378.4
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr3_-_170626376 | 0.21 |
ENST00000487522.1
ENST00000474417.1 |
EIF5A2
|
eukaryotic translation initiation factor 5A2 |
| chr12_+_112451222 | 0.21 |
ENST00000552052.1
|
ERP29
|
endoplasmic reticulum protein 29 |
| chr19_+_18530146 | 0.21 |
ENST00000348495.6
ENST00000270061.7 |
SSBP4
|
single stranded DNA binding protein 4 |
| chr19_-_19314162 | 0.21 |
ENST00000420605.3
ENST00000544883.1 ENST00000538165.2 ENST00000331552.7 |
NR2C2AP
|
nuclear receptor 2C2-associated protein |
| chr4_-_6565315 | 0.21 |
ENST00000506140.1
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr17_+_76210367 | 0.21 |
ENST00000592734.1
ENST00000587746.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr19_-_36499521 | 0.21 |
ENST00000397428.3
ENST00000503121.1 ENST00000340477.5 ENST00000324444.3 ENST00000490730.1 |
SYNE4
|
spectrin repeat containing, nuclear envelope family member 4 |
| chr19_-_23869970 | 0.21 |
ENST00000601010.1
|
ZNF675
|
zinc finger protein 675 |
| chr8_+_22446763 | 0.21 |
ENST00000450780.2
ENST00000430850.2 ENST00000447849.1 |
AC037459.4
|
Uncharacterized protein |
| chr22_-_20850128 | 0.21 |
ENST00000328879.4
|
KLHL22
|
kelch-like family member 22 |
| chr19_+_18043810 | 0.21 |
ENST00000445755.2
|
CCDC124
|
coiled-coil domain containing 124 |
| chr22_+_37415676 | 0.21 |
ENST00000401419.3
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr19_+_50321528 | 0.21 |
ENST00000312865.6
ENST00000595185.1 ENST00000538643.1 |
MED25
|
mediator complex subunit 25 |
| chr22_+_30821732 | 0.21 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr12_-_51418549 | 0.21 |
ENST00000548150.1
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr17_-_74533734 | 0.21 |
ENST00000589342.1
|
CYGB
|
cytoglobin |
| chr1_+_155099927 | 0.21 |
ENST00000368407.3
|
EFNA1
|
ephrin-A1 |
| chr19_-_21512115 | 0.21 |
ENST00000601295.1
|
ZNF708
|
zinc finger protein 708 |
| chr3_-_182880541 | 0.20 |
ENST00000470251.1
ENST00000265598.3 |
LAMP3
|
lysosomal-associated membrane protein 3 |
| chr13_-_21348050 | 0.20 |
ENST00000382754.4
|
N6AMT2
|
N-6 adenine-specific DNA methyltransferase 2 (putative) |
| chr21_-_46359760 | 0.20 |
ENST00000330551.3
ENST00000397841.1 ENST00000380070.4 |
C21orf67
|
chromosome 21 open reading frame 67 |
| chr14_+_31343747 | 0.20 |
ENST00000216361.4
ENST00000396618.3 ENST00000475087.1 |
COCH
|
cochlin |
| chr5_-_176738883 | 0.20 |
ENST00000513169.1
ENST00000423571.2 ENST00000502529.1 ENST00000427908.2 |
MXD3
|
MAX dimerization protein 3 |
| chr3_+_15469058 | 0.20 |
ENST00000432764.2
|
EAF1
|
ELL associated factor 1 |
| chr11_+_65383227 | 0.20 |
ENST00000355703.3
|
PCNXL3
|
pecanex-like 3 (Drosophila) |
| chr19_-_1592828 | 0.20 |
ENST00000592012.1
|
MBD3
|
methyl-CpG binding domain protein 3 |
| chr11_-_66725837 | 0.20 |
ENST00000393958.2
ENST00000393960.1 ENST00000524491.1 ENST00000355677.3 |
PC
|
pyruvate carboxylase |
| chr19_+_33668509 | 0.20 |
ENST00000592484.1
|
LRP3
|
low density lipoprotein receptor-related protein 3 |
| chr20_-_43743790 | 0.20 |
ENST00000307971.4
ENST00000372789.4 |
WFDC5
|
WAP four-disulfide core domain 5 |
| chr6_-_31648150 | 0.20 |
ENST00000375858.3
ENST00000383237.4 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr12_+_7022909 | 0.20 |
ENST00000537688.1
|
ENO2
|
enolase 2 (gamma, neuronal) |
| chr11_-_67771513 | 0.19 |
ENST00000227471.2
|
UNC93B1
|
unc-93 homolog B1 (C. elegans) |
| chr22_+_37415700 | 0.19 |
ENST00000397129.1
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr16_+_2098003 | 0.19 |
ENST00000439673.2
ENST00000350773.4 |
TSC2
|
tuberous sclerosis 2 |
| chr17_+_78075498 | 0.19 |
ENST00000302262.3
|
GAA
|
glucosidase, alpha; acid |
| chr6_-_26032288 | 0.19 |
ENST00000244661.2
|
HIST1H3B
|
histone cluster 1, H3b |
| chr19_+_8429031 | 0.19 |
ENST00000301455.2
ENST00000541807.1 ENST00000393962.2 |
ANGPTL4
|
angiopoietin-like 4 |
| chr19_-_13260992 | 0.19 |
ENST00000242770.5
ENST00000589083.1 ENST00000587230.1 |
STX10
|
syntaxin 10 |
| chr19_+_49436936 | 0.19 |
ENST00000221403.2
ENST00000523250.1 ENST00000522614.1 |
DHDH
|
dihydrodiol dehydrogenase (dimeric) |
| chr17_+_27047570 | 0.19 |
ENST00000472628.1
ENST00000578181.1 |
RPL23A
|
ribosomal protein L23a |
| chr22_-_19132154 | 0.19 |
ENST00000252137.6
|
DGCR14
|
DiGeorge syndrome critical region gene 14 |
| chr1_-_84543614 | 0.19 |
ENST00000605506.1
|
RP11-486G15.2
|
RP11-486G15.2 |
| chr1_-_45140227 | 0.19 |
ENST00000372237.3
|
TMEM53
|
transmembrane protein 53 |
| chr6_-_13290684 | 0.19 |
ENST00000606393.1
|
RP1-257A7.5
|
RP1-257A7.5 |
| chr17_-_43339453 | 0.19 |
ENST00000543122.1
|
SPATA32
|
spermatogenesis associated 32 |
| chr19_+_1040096 | 0.19 |
ENST00000263094.6
ENST00000524850.1 |
ABCA7
|
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr3_-_196695692 | 0.19 |
ENST00000412723.1
|
PIGZ
|
phosphatidylinositol glycan anchor biosynthesis, class Z |
| chr10_-_82116497 | 0.19 |
ENST00000372204.3
|
DYDC1
|
DPY30 domain containing 1 |
| chr19_-_59031118 | 0.18 |
ENST00000600990.1
|
ZBTB45
|
zinc finger and BTB domain containing 45 |
| chr14_-_100625932 | 0.18 |
ENST00000553834.1
|
DEGS2
|
delta(4)-desaturase, sphingolipid 2 |
| chr19_+_571277 | 0.18 |
ENST00000346916.4
ENST00000545507.2 |
BSG
|
basigin (Ok blood group) |
| chr1_-_45805607 | 0.18 |
ENST00000372104.1
ENST00000448481.1 ENST00000483127.1 ENST00000528013.2 ENST00000456914.2 |
MUTYH
|
mutY homolog |
| chr22_+_37415776 | 0.18 |
ENST00000341116.3
ENST00000429360.2 ENST00000404393.1 |
MPST
|
mercaptopyruvate sulfurtransferase |
| chr4_+_155548101 | 0.18 |
ENST00000502525.1
|
LRAT
|
lecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase) |
| chr17_+_40118773 | 0.18 |
ENST00000472031.1
|
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr17_+_40118805 | 0.18 |
ENST00000591072.1
ENST00000587679.1 ENST00000393888.1 ENST00000441615.2 |
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr11_-_615942 | 0.18 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr19_+_50879705 | 0.18 |
ENST00000598168.1
ENST00000411902.2 ENST00000253727.5 ENST00000597790.1 ENST00000597130.1 ENST00000599105.1 |
NR1H2
|
nuclear receptor subfamily 1, group H, member 2 |
| chr19_+_56159362 | 0.18 |
ENST00000593069.1
ENST00000308964.3 |
CCDC106
|
coiled-coil domain containing 106 |
| chr7_-_98805129 | 0.18 |
ENST00000327442.6
|
KPNA7
|
karyopherin alpha 7 (importin alpha 8) |
| chr22_+_24820341 | 0.18 |
ENST00000464977.1
ENST00000444262.2 |
ADORA2A
|
adenosine A2a receptor |
| chr22_-_20104700 | 0.18 |
ENST00000439169.2
ENST00000445045.1 ENST00000404751.3 ENST00000252136.7 ENST00000403707.3 |
TRMT2A
|
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
| chr1_-_52520828 | 0.18 |
ENST00000610127.1
|
TXNDC12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr22_-_36850991 | 0.18 |
ENST00000442579.1
|
RP5-1119A7.14
|
RP5-1119A7.14 |
| chr14_+_95078714 | 0.18 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr19_+_531713 | 0.18 |
ENST00000215574.4
|
CDC34
|
cell division cycle 34 |
| chr1_-_161039753 | 0.18 |
ENST00000368015.1
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr4_-_682960 | 0.18 |
ENST00000512249.1
ENST00000515118.1 ENST00000347950.5 |
MFSD7
|
major facilitator superfamily domain containing 7 |
| chr7_+_100271355 | 0.18 |
ENST00000436220.1
ENST00000424361.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr11_+_45944190 | 0.18 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr16_+_2079501 | 0.17 |
ENST00000563587.1
|
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr16_+_31119615 | 0.17 |
ENST00000394950.3
ENST00000287507.3 ENST00000219794.6 ENST00000561755.1 |
BCKDK
|
branched chain ketoacid dehydrogenase kinase |
| chr19_-_45909585 | 0.17 |
ENST00000593226.1
ENST00000418234.2 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chr1_-_50889155 | 0.17 |
ENST00000404795.3
|
DMRTA2
|
DMRT-like family A2 |
| chr17_+_42422662 | 0.17 |
ENST00000593167.1
ENST00000585512.1 ENST00000591740.1 ENST00000592783.1 ENST00000587387.1 ENST00000588237.1 ENST00000589265.1 |
GRN
|
granulin |
| chr19_-_5903714 | 0.17 |
ENST00000586349.1
ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12
NDUFA11
FUT5
|
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr22_-_21356375 | 0.17 |
ENST00000215742.4
ENST00000399133.2 |
THAP7
|
THAP domain containing 7 |
| chr19_+_2236509 | 0.17 |
ENST00000221494.5
|
SF3A2
|
splicing factor 3a, subunit 2, 66kDa |
| chr19_-_10230562 | 0.17 |
ENST00000587146.1
ENST00000588709.1 ENST00000253108.4 |
EIF3G
|
eukaryotic translation initiation factor 3, subunit G |
| chr1_-_11986442 | 0.17 |
ENST00000376572.3
ENST00000376576.3 |
KIAA2013
|
KIAA2013 |
| chr19_+_31658405 | 0.17 |
ENST00000589511.1
|
CTC-439O9.3
|
CTC-439O9.3 |
| chr2_-_20101701 | 0.17 |
ENST00000402414.1
ENST00000333610.3 |
TTC32
|
tetratricopeptide repeat domain 32 |
| chr7_-_105926058 | 0.17 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr1_-_228291136 | 0.17 |
ENST00000272139.4
|
C1orf35
|
chromosome 1 open reading frame 35 |
| chr16_+_81772633 | 0.17 |
ENST00000566191.1
ENST00000565272.1 ENST00000563954.1 ENST00000565054.1 |
RP11-960L18.1
PLCG2
|
RP11-960L18.1 phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| chr1_+_156698743 | 0.17 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chrX_+_149531606 | 0.16 |
ENST00000432680.2
|
MAMLD1
|
mastermind-like domain containing 1 |
| chr19_-_7167989 | 0.16 |
ENST00000600492.1
|
INSR
|
insulin receptor |
| chr15_+_75074915 | 0.16 |
ENST00000567123.1
ENST00000569462.1 |
CSK
|
c-src tyrosine kinase |
| chr7_+_76139741 | 0.16 |
ENST00000334348.3
ENST00000419923.2 ENST00000448265.3 ENST00000443097.2 |
UPK3B
|
uroplakin 3B |
| chr17_+_78075361 | 0.16 |
ENST00000577106.1
ENST00000390015.3 |
GAA
|
glucosidase, alpha; acid |
| chr16_+_30662085 | 0.16 |
ENST00000569864.1
|
PRR14
|
proline rich 14 |
| chr19_-_7698599 | 0.16 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr16_-_84220633 | 0.16 |
ENST00000566732.1
ENST00000561955.1 ENST00000564454.1 ENST00000341690.6 ENST00000541676.1 ENST00000570117.1 ENST00000564345.1 |
TAF1C
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa |
| chr19_-_10491234 | 0.16 |
ENST00000524462.1
ENST00000531836.1 ENST00000525621.1 |
TYK2
|
tyrosine kinase 2 |
| chr15_-_65809625 | 0.16 |
ENST00000560436.1
|
DPP8
|
dipeptidyl-peptidase 8 |
| chr22_-_31503490 | 0.16 |
ENST00000400299.2
|
SELM
|
Selenoprotein M |
| chr19_-_18197799 | 0.16 |
ENST00000430026.3
ENST00000593993.2 |
IL12RB1
|
interleukin 12 receptor, beta 1 |
| chr12_-_8380183 | 0.16 |
ENST00000442295.2
ENST00000307435.6 ENST00000538603.1 |
FAM90A1
|
family with sequence similarity 90, member A1 |
| chr9_-_136933615 | 0.16 |
ENST00000371834.2
|
BRD3
|
bromodomain containing 3 |
| chr22_-_20850070 | 0.16 |
ENST00000440659.2
ENST00000458248.1 ENST00000443285.1 ENST00000444967.1 ENST00000451553.1 ENST00000431430.1 |
KLHL22
|
kelch-like family member 22 |
| chr19_+_3136115 | 0.16 |
ENST00000262958.3
|
GNA15
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class) |
| chr2_-_70475730 | 0.16 |
ENST00000445587.1
ENST00000433529.2 ENST00000415783.2 |
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr17_-_42402138 | 0.16 |
ENST00000592857.1
ENST00000586016.1 ENST00000590194.1 ENST00000377095.5 ENST00000588049.1 ENST00000586633.1 ENST00000537904.2 ENST00000585636.1 ENST00000585523.1 ENST00000225308.8 |
SLC25A39
|
solute carrier family 25, member 39 |
| chr3_+_52448539 | 0.16 |
ENST00000461861.1
|
PHF7
|
PHD finger protein 7 |
| chr19_+_56159421 | 0.16 |
ENST00000587213.1
|
CCDC106
|
coiled-coil domain containing 106 |
| chr11_+_65687158 | 0.16 |
ENST00000532933.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr18_-_53303123 | 0.16 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr19_+_40873617 | 0.16 |
ENST00000599353.1
|
PLD3
|
phospholipase D family, member 3 |
| chr11_-_18270182 | 0.16 |
ENST00000528349.1
ENST00000526900.1 ENST00000529528.1 ENST00000414546.2 ENST00000256733.4 |
SAA2
|
serum amyloid A2 |
| chr16_-_5083917 | 0.15 |
ENST00000312251.3
ENST00000381955.3 |
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr11_+_117049445 | 0.15 |
ENST00000324225.4
ENST00000532960.1 |
SIDT2
|
SID1 transmembrane family, member 2 |
| chr22_+_21400229 | 0.15 |
ENST00000342608.4
ENST00000543388.1 ENST00000442047.1 |
AC002472.13
|
Leucine-rich repeat-containing protein LOC400891 |
| chr19_+_17186577 | 0.15 |
ENST00000595618.1
ENST00000594824.1 |
MYO9B
|
myosin IXB |
| chr19_-_47735918 | 0.15 |
ENST00000449228.1
ENST00000300880.7 ENST00000341983.4 |
BBC3
|
BCL2 binding component 3 |
| chr16_+_57126428 | 0.15 |
ENST00000290776.8
|
CPNE2
|
copine II |
| chr19_+_11457232 | 0.15 |
ENST00000587531.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr16_-_84150392 | 0.15 |
ENST00000570012.1
|
MBTPS1
|
membrane-bound transcription factor peptidase, site 1 |
| chr4_-_1242764 | 0.15 |
ENST00000514210.1
ENST00000382952.3 ENST00000290921.6 |
CTBP1
|
C-terminal binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF524
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.5 | 4.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 1.2 | GO:0009440 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.2 | 0.5 | GO:1901076 | positive regulation of engulfment of apoptotic cell(GO:1901076) |
| 0.1 | 0.6 | GO:0006562 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.3 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.1 | 0.3 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.3 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.9 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 0.4 | GO:1904806 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.1 | 0.3 | GO:1901207 | regulation of heart looping(GO:1901207) positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.1 | 0.3 | GO:1903939 | regulation of TORC2 signaling(GO:1903939) |
| 0.1 | 0.2 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.1 | 0.9 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.2 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.1 | 0.2 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 | 0.4 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 0.1 | GO:0010985 | negative regulation of lipoprotein particle clearance(GO:0010985) |
| 0.1 | 0.2 | GO:0090187 | positive regulation of high-density lipoprotein particle assembly(GO:0090108) positive regulation of pancreatic juice secretion(GO:0090187) positive regulation of secretion of lysosomal enzymes(GO:0090340) |
| 0.1 | 0.3 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 0.4 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.2 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.2 | GO:0030451 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.1 | 0.2 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 0.2 | GO:0090410 | malonate catabolic process(GO:0090410) |
| 0.1 | 0.2 | GO:2001178 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.1 | 0.3 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.2 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.3 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.1 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.5 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.2 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.0 | 0.2 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.3 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.0 | 0.2 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:1904351 | negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.0 | 0.1 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.0 | 0.1 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.0 | 0.2 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.2 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.4 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.1 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.2 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.1 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.2 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.0 | 0.1 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.0 | 0.2 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.3 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.5 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.0 | 0.2 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.1 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.7 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.0 | 0.1 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.1 | GO:0071106 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.0 | 0.3 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0003068 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.3 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.2 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.2 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) |
| 0.0 | 0.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.4 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.2 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.2 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) RNA repair(GO:0042245) |
| 0.0 | 0.0 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.2 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.1 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.1 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:1900241 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.1 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.1 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.1 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.0 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.2 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) L-kynurenine metabolic process(GO:0097052) |
| 0.0 | 0.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.1 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.1 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 0.1 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.0 | 0.1 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.0 | GO:0001825 | blastocyst formation(GO:0001825) trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.0 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.1 | GO:1901859 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.0 | 0.1 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.0 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.0 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.0 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:0033123 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.1 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.2 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.4 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.0 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.0 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.1 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
| 0.0 | 0.1 | GO:0038112 | interleukin-8-mediated signaling pathway(GO:0038112) |
| 0.0 | 0.0 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.1 | GO:0060482 | lobar bronchus development(GO:0060482) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.0 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.2 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.0 | GO:1902823 | negative regulation of late endosome to lysosome transport(GO:1902823) |
| 0.0 | 0.0 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.0 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 0.0 | GO:0048242 | regulation of epinephrine secretion(GO:0014060) negative regulation of epinephrine secretion(GO:0032811) epinephrine secretion(GO:0048242) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.0 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.0 | GO:0060829 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.2 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.0 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.0 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.2 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) G1 DNA damage checkpoint(GO:0044783) mitotic G1/S transition checkpoint(GO:0044819) |
| 0.0 | 0.1 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.0 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.0 | 0.1 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.1 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.2 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.2 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.1 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.5 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.2 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.1 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.1 | GO:0042022 | interleukin-12 receptor complex(GO:0042022) |
| 0.0 | 0.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.0 | 0.4 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.5 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.1 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.5 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 4.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.0 | 0.2 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0031310 | intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.6 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.4 | GO:0044453 | nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.0 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) |
| 0.0 | 0.0 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 0.8 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.2 | 0.4 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.2 | 0.6 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.1 | 0.5 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.5 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.1 | 0.3 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.3 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.1 | 0.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.5 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 0.2 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.2 | GO:0032406 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.1 | 0.2 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.1 | 0.2 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.1 | 0.2 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 0.2 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.2 | GO:0090409 | malonyl-CoA synthetase activity(GO:0090409) |
| 0.1 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.2 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.3 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.2 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 0.6 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0016517 | interleukin-12 receptor activity(GO:0016517) |
| 0.0 | 0.1 | GO:0047536 | 2-aminoadipate transaminase activity(GO:0047536) |
| 0.0 | 0.1 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.3 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.2 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.2 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.2 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.2 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0052835 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.1 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.0 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.3 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.3 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.3 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.2 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0016416 | O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.4 | GO:0001164 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.0 | GO:0031071 | cysteine desulfurase activity(GO:0031071) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.0 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.1 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.0 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.0 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.0 | 0.2 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.0 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.0 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.0 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.3 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.0 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.6 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.1 | REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | Genes involved in Gastrin-CREB signalling pathway via PKC and MAPK |
| 0.0 | 0.8 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.0 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.1 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |