Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
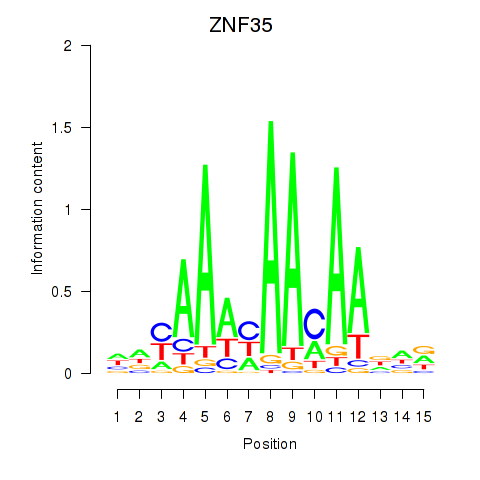
Results for ZNF35
Z-value: 0.47
Transcription factors associated with ZNF35
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF35
|
ENSG00000169981.6 | zinc finger protein 35 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF35 | hg19_v2_chr3_+_44690211_44690267 | 0.00 | 1.0e+00 | Click! |
Activity profile of ZNF35 motif
Sorted Z-values of ZNF35 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_35553045 | 0.27 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr4_+_75174180 | 0.23 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chr14_+_76776957 | 0.23 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr9_+_42671887 | 0.21 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr12_+_51318513 | 0.19 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr12_+_25205155 | 0.18 |
ENST00000550945.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr5_-_68339648 | 0.18 |
ENST00000479830.2
|
CTC-498J12.1
|
CTC-498J12.1 |
| chr21_+_17792672 | 0.17 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr12_+_25205446 | 0.17 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr3_-_171489085 | 0.16 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chr5_-_39203093 | 0.16 |
ENST00000515010.1
|
FYB
|
FYN binding protein |
| chr13_-_41240717 | 0.14 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr4_-_89442940 | 0.13 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr3_+_29323043 | 0.13 |
ENST00000452462.1
ENST00000456853.1 |
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr12_+_25205666 | 0.13 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr17_-_58042075 | 0.13 |
ENST00000305783.8
ENST00000589113.1 ENST00000442346.2 |
RNFT1
|
ring finger protein, transmembrane 1 |
| chr5_+_139027877 | 0.13 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr1_+_61869748 | 0.11 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr14_-_92198403 | 0.10 |
ENST00000553329.1
ENST00000256343.3 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chrX_-_138724677 | 0.10 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr1_-_243349684 | 0.09 |
ENST00000522895.1
|
CEP170
|
centrosomal protein 170kDa |
| chr15_+_96876340 | 0.09 |
ENST00000453270.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr10_-_4285923 | 0.09 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr3_-_148939835 | 0.09 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr3_-_49967292 | 0.08 |
ENST00000455683.2
|
MON1A
|
MON1 secretory trafficking family member A |
| chr6_-_135271219 | 0.07 |
ENST00000367847.2
ENST00000367845.2 |
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr2_+_177015122 | 0.07 |
ENST00000468418.3
|
HOXD3
|
homeobox D3 |
| chr5_+_139028510 | 0.07 |
ENST00000502336.1
ENST00000520967.1 ENST00000511048.1 |
CXXC5
|
CXXC finger protein 5 |
| chr6_+_32812568 | 0.07 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr8_-_101157680 | 0.06 |
ENST00000428847.2
|
FBXO43
|
F-box protein 43 |
| chr8_-_145754428 | 0.05 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr15_-_76352069 | 0.05 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr1_-_161008697 | 0.05 |
ENST00000318289.10
ENST00000368023.3 ENST00000368024.1 ENST00000423014.2 |
TSTD1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr21_+_35552978 | 0.04 |
ENST00000428914.2
ENST00000609062.1 ENST00000609947.1 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr13_+_109248500 | 0.04 |
ENST00000356711.2
|
MYO16
|
myosin XVI |
| chr12_-_22487588 | 0.04 |
ENST00000381424.3
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr16_+_21623958 | 0.04 |
ENST00000568826.1
|
METTL9
|
methyltransferase like 9 |
| chr1_-_169599353 | 0.04 |
ENST00000367793.2
ENST00000367794.2 ENST00000367792.2 ENST00000367791.2 ENST00000367788.2 |
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62) |
| chr6_+_76599809 | 0.04 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chrM_+_4431 | 0.03 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr3_+_69985734 | 0.03 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr12_-_22487618 | 0.03 |
ENST00000404299.3
ENST00000396037.4 |
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr1_-_12677714 | 0.03 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr7_+_74072288 | 0.03 |
ENST00000443166.1
|
GTF2I
|
general transcription factor IIi |
| chr1_+_220701456 | 0.02 |
ENST00000366918.4
ENST00000402574.1 |
MARK1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr4_-_89744365 | 0.02 |
ENST00000513837.1
ENST00000503556.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr7_+_80231466 | 0.02 |
ENST00000309881.7
ENST00000534394.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr4_+_129730947 | 0.02 |
ENST00000452328.2
ENST00000504089.1 |
PHF17
|
jade family PHD finger 1 |
| chr10_-_52645416 | 0.02 |
ENST00000374001.2
ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr4_-_99850243 | 0.02 |
ENST00000280892.6
ENST00000511644.1 ENST00000504432.1 ENST00000505992.1 |
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr13_-_47471155 | 0.02 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr7_-_83824449 | 0.01 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr3_+_10157276 | 0.01 |
ENST00000530758.1
ENST00000256463.6 |
BRK1
|
BRICK1, SCAR/WAVE actin-nucleating complex subunit |
| chr12_+_4382917 | 0.01 |
ENST00000261254.3
|
CCND2
|
cyclin D2 |
| chr18_+_39535239 | 0.01 |
ENST00000585528.1
|
PIK3C3
|
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr2_+_166150541 | 0.01 |
ENST00000283256.6
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr2_-_230787879 | 0.01 |
ENST00000435716.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr3_+_141106458 | 0.01 |
ENST00000509883.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr5_-_58571935 | 0.01 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr8_-_116680833 | 0.01 |
ENST00000220888.5
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr7_+_123488124 | 0.00 |
ENST00000476325.1
|
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr13_-_36429763 | 0.00 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF35
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |