Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
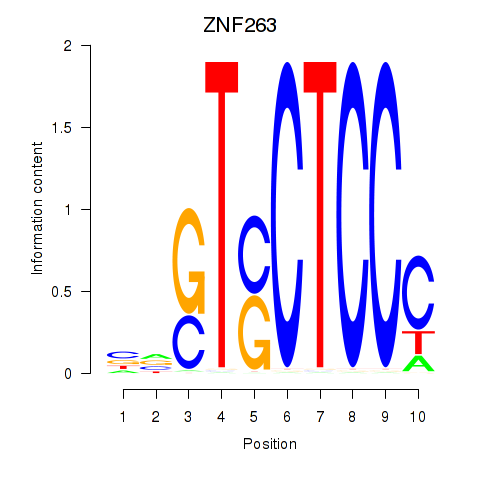
Results for ZNF263
Z-value: 0.74
Transcription factors associated with ZNF263
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF263
|
ENSG00000006194.6 | zinc finger protein 263 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF263 | hg19_v2_chr16_+_3333443_3333545 | -0.71 | 1.2e-01 | Click! |
Activity profile of ZNF263 motif
Sorted Z-values of ZNF263 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_125932685 | 0.35 |
ENST00000527967.1
|
CDON
|
cell adhesion associated, oncogene regulated |
| chr19_-_35626104 | 0.27 |
ENST00000310123.3
ENST00000392225.3 |
LGI4
|
leucine-rich repeat LGI family, member 4 |
| chr11_-_46142615 | 0.26 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr19_-_55658687 | 0.26 |
ENST00000593046.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr6_-_153452356 | 0.25 |
ENST00000206262.1
|
RGS17
|
regulator of G-protein signaling 17 |
| chr4_+_26859300 | 0.25 |
ENST00000494628.2
|
STIM2
|
stromal interaction molecule 2 |
| chr11_+_64004888 | 0.24 |
ENST00000541681.1
|
VEGFB
|
vascular endothelial growth factor B |
| chr15_+_91427642 | 0.24 |
ENST00000328850.3
ENST00000414248.2 |
FES
|
feline sarcoma oncogene |
| chr2_+_27309605 | 0.23 |
ENST00000260599.6
ENST00000260598.5 ENST00000429697.1 |
KHK
|
ketohexokinase (fructokinase) |
| chr9_+_84304628 | 0.23 |
ENST00000437181.1
|
RP11-154D17.1
|
RP11-154D17.1 |
| chr16_+_532503 | 0.22 |
ENST00000412256.1
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II) |
| chr12_+_6930703 | 0.21 |
ENST00000311268.3
|
GPR162
|
G protein-coupled receptor 162 |
| chr1_+_211432700 | 0.21 |
ENST00000452621.2
|
RCOR3
|
REST corepressor 3 |
| chr3_+_167453026 | 0.20 |
ENST00000472941.1
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr1_-_40782347 | 0.20 |
ENST00000417105.1
|
COL9A2
|
collagen, type IX, alpha 2 |
| chr2_+_86116396 | 0.20 |
ENST00000455121.3
|
AC105053.4
|
AC105053.4 |
| chr15_-_90358564 | 0.20 |
ENST00000559874.1
|
ANPEP
|
alanyl (membrane) aminopeptidase |
| chr11_-_64949305 | 0.20 |
ENST00000526623.1
|
AP003068.23
|
Uncharacterized protein |
| chr17_+_36508826 | 0.20 |
ENST00000580660.1
|
SOCS7
|
suppressor of cytokine signaling 7 |
| chr11_-_46142505 | 0.20 |
ENST00000524497.1
ENST00000418153.2 |
PHF21A
|
PHD finger protein 21A |
| chr11_-_46142948 | 0.19 |
ENST00000257821.4
|
PHF21A
|
PHD finger protein 21A |
| chrX_+_107068959 | 0.19 |
ENST00000451923.1
|
MID2
|
midline 2 |
| chr1_-_85156417 | 0.18 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr15_-_71146347 | 0.18 |
ENST00000559140.2
|
LARP6
|
La ribonucleoprotein domain family, member 6 |
| chr16_-_67427389 | 0.18 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr20_-_55841662 | 0.18 |
ENST00000395863.3
ENST00000450594.2 |
BMP7
|
bone morphogenetic protein 7 |
| chr19_-_47975417 | 0.18 |
ENST00000236877.6
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr1_+_840205 | 0.18 |
ENST00000607769.1
|
RP11-54O7.16
|
RP11-54O7.16 |
| chr14_-_27066960 | 0.18 |
ENST00000539517.2
|
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr11_+_64053311 | 0.18 |
ENST00000540370.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr19_-_47975106 | 0.17 |
ENST00000539381.1
ENST00000594353.1 ENST00000542837.1 |
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chrX_-_41782592 | 0.17 |
ENST00000378158.1
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr9_+_86595626 | 0.17 |
ENST00000445877.1
ENST00000325875.3 |
RMI1
|
RecQ mediated genome instability 1 |
| chr7_-_105516923 | 0.17 |
ENST00000478915.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr19_-_10679697 | 0.17 |
ENST00000335766.2
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr11_-_129872672 | 0.16 |
ENST00000531431.1
ENST00000527581.1 |
PRDM10
|
PR domain containing 10 |
| chr3_-_50605150 | 0.16 |
ENST00000357203.3
|
C3orf18
|
chromosome 3 open reading frame 18 |
| chr19_-_10679644 | 0.16 |
ENST00000393599.2
|
CDKN2D
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr1_-_41950342 | 0.16 |
ENST00000372587.4
|
EDN2
|
endothelin 2 |
| chr14_+_77564701 | 0.16 |
ENST00000557115.1
|
KIAA1737
|
CLOCK-interacting pacemaker |
| chr17_+_7461580 | 0.16 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr10_-_77161004 | 0.15 |
ENST00000418818.2
|
RP11-399K21.11
|
RP11-399K21.11 |
| chr14_-_102605983 | 0.15 |
ENST00000334701.7
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr4_+_184365841 | 0.15 |
ENST00000510928.1
|
CDKN2AIP
|
CDKN2A interacting protein |
| chr14_+_75746664 | 0.15 |
ENST00000557139.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chrX_-_41782683 | 0.15 |
ENST00000378163.1
ENST00000378154.1 |
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr19_-_38714847 | 0.14 |
ENST00000420980.2
ENST00000355526.4 |
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr2_+_204193101 | 0.14 |
ENST00000430418.1
ENST00000424558.1 ENST00000261016.6 |
ABI2
|
abl-interactor 2 |
| chr20_-_55841398 | 0.14 |
ENST00000395864.3
|
BMP7
|
bone morphogenetic protein 7 |
| chr3_-_185826718 | 0.14 |
ENST00000413301.1
ENST00000421809.1 |
ETV5
|
ets variant 5 |
| chr20_-_44455976 | 0.14 |
ENST00000372555.3
|
TNNC2
|
troponin C type 2 (fast) |
| chr11_-_71752838 | 0.14 |
ENST00000537930.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr15_+_100347228 | 0.14 |
ENST00000559714.1
ENST00000560059.1 |
CTD-2054N24.2
|
Uncharacterized protein |
| chr4_-_185395191 | 0.14 |
ENST00000510814.1
ENST00000507523.1 ENST00000506230.1 |
IRF2
|
interferon regulatory factor 2 |
| chr2_+_48757278 | 0.14 |
ENST00000404752.1
ENST00000406226.1 |
STON1
|
stonin 1 |
| chr19_-_47975143 | 0.13 |
ENST00000597014.1
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr9_-_130213652 | 0.13 |
ENST00000536368.1
ENST00000361436.5 |
RPL12
|
ribosomal protein L12 |
| chr19_+_57106624 | 0.13 |
ENST00000599599.1
|
ZNF71
|
zinc finger protein 71 |
| chr19_-_49565254 | 0.13 |
ENST00000593537.1
|
NTF4
|
neurotrophin 4 |
| chr1_-_2461684 | 0.13 |
ENST00000378453.3
|
HES5
|
hes family bHLH transcription factor 5 |
| chrX_+_107069063 | 0.13 |
ENST00000262843.6
|
MID2
|
midline 2 |
| chr11_-_71752571 | 0.13 |
ENST00000544238.1
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr17_+_72733350 | 0.13 |
ENST00000392613.5
ENST00000392612.3 ENST00000392610.1 |
RAB37
|
RAB37, member RAS oncogene family |
| chr8_+_98656693 | 0.13 |
ENST00000519934.1
|
MTDH
|
metadherin |
| chr2_-_74645669 | 0.13 |
ENST00000518401.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr12_-_57030096 | 0.13 |
ENST00000549506.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr13_-_46626820 | 0.13 |
ENST00000428921.1
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr2_-_158732340 | 0.13 |
ENST00000539637.1
ENST00000413751.1 ENST00000434821.1 ENST00000424669.1 |
ACVR1
|
activin A receptor, type I |
| chr1_+_145525015 | 0.13 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chr22_-_36424458 | 0.13 |
ENST00000438146.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr2_-_174828892 | 0.13 |
ENST00000418194.2
|
SP3
|
Sp3 transcription factor |
| chr17_-_6543973 | 0.13 |
ENST00000571642.1
ENST00000572370.1 |
KIAA0753
|
KIAA0753 |
| chr15_-_71146480 | 0.12 |
ENST00000299213.8
|
LARP6
|
La ribonucleoprotein domain family, member 6 |
| chr5_-_81046841 | 0.12 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr16_-_31021717 | 0.12 |
ENST00000565419.1
|
STX1B
|
syntaxin 1B |
| chr5_+_154135453 | 0.12 |
ENST00000517616.1
ENST00000518892.1 |
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr21_-_40720995 | 0.12 |
ENST00000380749.5
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr19_-_31840438 | 0.12 |
ENST00000240587.4
|
TSHZ3
|
teashirt zinc finger homeobox 3 |
| chr4_-_113207048 | 0.12 |
ENST00000361717.3
|
TIFA
|
TRAF-interacting protein with forkhead-associated domain |
| chr1_+_209859510 | 0.12 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr19_+_496454 | 0.12 |
ENST00000346144.4
ENST00000215637.3 ENST00000382683.4 |
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr12_-_96793142 | 0.12 |
ENST00000552262.1
ENST00000551816.1 ENST00000552496.1 |
CDK17
|
cyclin-dependent kinase 17 |
| chr15_+_85427879 | 0.12 |
ENST00000338602.2
ENST00000537216.1 ENST00000538177.1 ENST00000537624.1 |
SLC28A1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr19_-_56092187 | 0.12 |
ENST00000325421.4
ENST00000592239.1 |
ZNF579
|
zinc finger protein 579 |
| chr1_+_205682497 | 0.12 |
ENST00000598338.1
|
AC119673.1
|
AC119673.1 |
| chr15_-_57025728 | 0.12 |
ENST00000559352.1
|
ZNF280D
|
zinc finger protein 280D |
| chr15_+_91427691 | 0.12 |
ENST00000559355.1
ENST00000394302.1 |
FES
|
feline sarcoma oncogene |
| chr3_+_157823609 | 0.12 |
ENST00000480820.1
|
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr5_+_5140436 | 0.12 |
ENST00000511368.1
ENST00000274181.7 |
ADAMTS16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16 |
| chr1_+_212782012 | 0.12 |
ENST00000341491.4
ENST00000366985.1 |
ATF3
|
activating transcription factor 3 |
| chr17_+_68165657 | 0.11 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr11_-_7041466 | 0.11 |
ENST00000536068.1
ENST00000278314.4 |
ZNF214
|
zinc finger protein 214 |
| chr1_+_164528437 | 0.11 |
ENST00000485769.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr7_-_73153178 | 0.11 |
ENST00000437775.2
ENST00000222800.3 |
ABHD11
|
abhydrolase domain containing 11 |
| chr12_-_124456598 | 0.11 |
ENST00000539761.1
ENST00000539551.1 |
CCDC92
|
coiled-coil domain containing 92 |
| chr6_+_32821924 | 0.11 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr19_-_2944907 | 0.11 |
ENST00000314531.4
|
ZNF77
|
zinc finger protein 77 |
| chr11_-_72852958 | 0.11 |
ENST00000458644.2
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chrX_-_47509994 | 0.11 |
ENST00000343894.4
|
ELK1
|
ELK1, member of ETS oncogene family |
| chrX_-_55187588 | 0.11 |
ENST00000472571.2
ENST00000332132.4 ENST00000425133.2 ENST00000358460.4 |
FAM104B
|
family with sequence similarity 104, member B |
| chr8_-_42751820 | 0.11 |
ENST00000526349.1
ENST00000527424.1 ENST00000534961.1 ENST00000319073.4 |
RNF170
|
ring finger protein 170 |
| chr8_-_26371608 | 0.11 |
ENST00000522362.2
|
PNMA2
|
paraneoplastic Ma antigen 2 |
| chr8_+_26435359 | 0.11 |
ENST00000311151.5
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr2_-_224702740 | 0.11 |
ENST00000444408.1
|
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit |
| chr19_-_51845378 | 0.11 |
ENST00000335624.4
|
VSIG10L
|
V-set and immunoglobulin domain containing 10 like |
| chr3_-_182703688 | 0.11 |
ENST00000466812.1
ENST00000487822.1 ENST00000460412.1 ENST00000469954.1 |
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr12_+_6930964 | 0.11 |
ENST00000382315.3
|
GPR162
|
G protein-coupled receptor 162 |
| chr15_-_77712429 | 0.11 |
ENST00000564328.1
ENST00000558305.1 |
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr11_+_33061336 | 0.11 |
ENST00000602733.1
|
TCP11L1
|
t-complex 11, testis-specific-like 1 |
| chr2_-_27579623 | 0.11 |
ENST00000457748.1
|
GTF3C2
|
general transcription factor IIIC, polypeptide 2, beta 110kDa |
| chr22_+_17082732 | 0.11 |
ENST00000558085.2
ENST00000592918.1 ENST00000400593.2 ENST00000592107.1 ENST00000426585.1 ENST00000591299.1 |
TPTEP1
|
transmembrane phosphatase with tensin homology pseudogene 1 |
| chr12_+_66218903 | 0.11 |
ENST00000393577.3
|
HMGA2
|
high mobility group AT-hook 2 |
| chr19_+_37960466 | 0.11 |
ENST00000589725.1
|
ZNF570
|
zinc finger protein 570 |
| chr17_+_62503073 | 0.11 |
ENST00000580188.1
ENST00000581056.1 |
CEP95
|
centrosomal protein 95kDa |
| chr4_+_56262115 | 0.11 |
ENST00000506198.1
ENST00000381334.5 ENST00000542052.1 |
TMEM165
|
transmembrane protein 165 |
| chr8_+_26240666 | 0.11 |
ENST00000523949.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr1_-_23694794 | 0.11 |
ENST00000374608.3
|
ZNF436
|
zinc finger protein 436 |
| chr2_+_65663812 | 0.11 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr18_-_70535177 | 0.10 |
ENST00000327305.6
|
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr10_+_76586348 | 0.10 |
ENST00000372724.1
ENST00000287239.4 ENST00000372714.1 |
KAT6B
|
K(lysine) acetyltransferase 6B |
| chr9_+_8858102 | 0.10 |
ENST00000447950.1
ENST00000430766.1 |
RP11-75C9.1
|
RP11-75C9.1 |
| chr14_+_102606181 | 0.10 |
ENST00000335263.5
ENST00000322340.5 ENST00000424963.2 ENST00000342702.3 ENST00000556807.1 ENST00000499851.2 ENST00000558567.1 ENST00000299135.6 ENST00000454394.2 ENST00000556511.2 |
WDR20
|
WD repeat domain 20 |
| chr14_+_62331592 | 0.10 |
ENST00000554436.1
|
CTD-2277K2.1
|
CTD-2277K2.1 |
| chr1_+_14075903 | 0.10 |
ENST00000343137.4
ENST00000503842.1 ENST00000407521.3 ENST00000505823.1 |
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr11_+_66115304 | 0.10 |
ENST00000531602.1
|
RP11-867G23.8
|
Uncharacterized protein |
| chr19_-_31840130 | 0.10 |
ENST00000558569.1
|
TSHZ3
|
teashirt zinc finger homeobox 3 |
| chr3_+_38323785 | 0.10 |
ENST00000466887.1
ENST00000448498.1 |
SLC22A14
|
solute carrier family 22, member 14 |
| chr13_-_46626847 | 0.10 |
ENST00000242848.4
ENST00000282007.3 |
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr8_+_26434578 | 0.10 |
ENST00000493789.2
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr17_+_4643337 | 0.10 |
ENST00000592813.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr6_-_111136299 | 0.10 |
ENST00000457688.1
|
CDK19
|
cyclin-dependent kinase 19 |
| chr12_-_124457163 | 0.10 |
ENST00000535556.1
|
CCDC92
|
coiled-coil domain containing 92 |
| chr15_-_71146460 | 0.10 |
ENST00000344870.4
|
LARP6
|
La ribonucleoprotein domain family, member 6 |
| chr5_+_61602236 | 0.10 |
ENST00000514082.1
ENST00000407818.3 |
KIF2A
|
kinesin heavy chain member 2A |
| chr6_-_31514516 | 0.10 |
ENST00000303892.5
ENST00000483251.1 |
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr1_-_21113765 | 0.10 |
ENST00000438032.1
ENST00000424732.1 ENST00000437575.1 |
HP1BP3
|
heterochromatin protein 1, binding protein 3 |
| chr3_+_184097836 | 0.10 |
ENST00000204604.1
ENST00000310236.3 |
CHRD
|
chordin |
| chr1_+_224803995 | 0.10 |
ENST00000272133.3
|
CNIH3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr12_+_96588279 | 0.10 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr21_-_40720974 | 0.10 |
ENST00000380748.1
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr7_-_151574191 | 0.10 |
ENST00000287878.4
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr11_+_125034586 | 0.10 |
ENST00000298282.9
|
PKNOX2
|
PBX/knotted 1 homeobox 2 |
| chr5_-_81046922 | 0.10 |
ENST00000514493.1
ENST00000320672.4 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr13_-_33002151 | 0.10 |
ENST00000495479.1
ENST00000343281.4 ENST00000464470.1 ENST00000380139.4 ENST00000380133.2 |
N4BP2L1
|
NEDD4 binding protein 2-like 1 |
| chr9_-_3525968 | 0.10 |
ENST00000382004.3
ENST00000302303.1 ENST00000449190.1 |
RFX3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr9_-_8857776 | 0.10 |
ENST00000481079.1
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D |
| chr21_-_43916433 | 0.10 |
ENST00000291536.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr19_-_51071302 | 0.10 |
ENST00000389201.3
ENST00000600381.1 |
LRRC4B
|
leucine rich repeat containing 4B |
| chr13_+_98628886 | 0.10 |
ENST00000490680.1
ENST00000539640.1 ENST00000403772.3 |
IPO5
|
importin 5 |
| chr12_+_57984965 | 0.10 |
ENST00000540759.2
ENST00000551772.1 ENST00000550465.1 ENST00000354947.5 |
PIP4K2C
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr17_+_8339340 | 0.10 |
ENST00000580012.1
|
NDEL1
|
nudE neurodevelopment protein 1-like 1 |
| chr1_-_226374373 | 0.09 |
ENST00000366812.5
|
ACBD3
|
acyl-CoA binding domain containing 3 |
| chr11_-_47521309 | 0.09 |
ENST00000535982.1
|
CELF1
|
CUGBP, Elav-like family member 1 |
| chr6_+_32938692 | 0.09 |
ENST00000443797.2
|
BRD2
|
bromodomain containing 2 |
| chr2_-_11605966 | 0.09 |
ENST00000307236.4
ENST00000542100.1 ENST00000546212.1 |
E2F6
|
E2F transcription factor 6 |
| chr7_+_100728720 | 0.09 |
ENST00000306085.6
ENST00000412507.1 |
TRIM56
|
tripartite motif containing 56 |
| chr8_+_61429728 | 0.09 |
ENST00000529579.1
|
RAB2A
|
RAB2A, member RAS oncogene family |
| chr20_+_13202418 | 0.09 |
ENST00000262487.4
|
ISM1
|
isthmin 1, angiogenesis inhibitor |
| chr3_+_155588375 | 0.09 |
ENST00000295920.7
|
GMPS
|
guanine monphosphate synthase |
| chr4_+_15471489 | 0.09 |
ENST00000424120.1
ENST00000413206.1 ENST00000438599.2 ENST00000511544.1 ENST00000512702.1 ENST00000507954.1 ENST00000515124.1 ENST00000503292.1 ENST00000503658.1 |
CC2D2A
|
coiled-coil and C2 domain containing 2A |
| chr19_-_12163754 | 0.09 |
ENST00000547628.1
|
ZNF878
|
zinc finger protein 878 |
| chrX_-_150067069 | 0.09 |
ENST00000466436.1
|
CD99L2
|
CD99 molecule-like 2 |
| chr11_-_47546250 | 0.09 |
ENST00000543178.1
|
CELF1
|
CUGBP, Elav-like family member 1 |
| chr4_-_69083720 | 0.09 |
ENST00000432593.3
|
TMPRSS11BNL
|
TMPRSS11B N-terminal like |
| chr11_-_6341844 | 0.09 |
ENST00000303927.3
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chrX_+_69674943 | 0.09 |
ENST00000542398.1
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr2_+_204192942 | 0.09 |
ENST00000295851.5
ENST00000261017.5 |
ABI2
|
abl-interactor 2 |
| chr15_+_49462397 | 0.09 |
ENST00000396509.2
|
GALK2
|
galactokinase 2 |
| chr13_-_52026730 | 0.09 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr6_+_35773070 | 0.09 |
ENST00000373853.1
ENST00000360215.1 |
LHFPL5
|
lipoma HMGIC fusion partner-like 5 |
| chr11_-_71639670 | 0.09 |
ENST00000533047.1
ENST00000529844.1 |
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr15_-_57210769 | 0.09 |
ENST00000559000.1
|
ZNF280D
|
zinc finger protein 280D |
| chr8_+_31496809 | 0.09 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chr19_+_50433476 | 0.09 |
ENST00000596658.1
|
ATF5
|
activating transcription factor 5 |
| chr6_-_33714667 | 0.09 |
ENST00000293756.4
|
IP6K3
|
inositol hexakisphosphate kinase 3 |
| chr8_+_70379072 | 0.09 |
ENST00000529134.1
|
SULF1
|
sulfatase 1 |
| chr3_-_193272741 | 0.09 |
ENST00000392443.3
|
ATP13A4
|
ATPase type 13A4 |
| chr5_+_10564432 | 0.09 |
ENST00000296657.5
|
ANKRD33B
|
ankyrin repeat domain 33B |
| chr14_+_77564440 | 0.09 |
ENST00000361786.2
ENST00000555437.1 ENST00000555611.1 ENST00000554658.1 |
KIAA1737
|
CLOCK-interacting pacemaker |
| chr1_-_92351666 | 0.09 |
ENST00000465892.2
ENST00000417833.2 |
TGFBR3
|
transforming growth factor, beta receptor III |
| chr1_+_145524891 | 0.09 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chr9_+_123884038 | 0.09 |
ENST00000373847.1
|
CNTRL
|
centriolin |
| chr11_+_29181503 | 0.09 |
ENST00000530960.1
|
RP11-466I1.1
|
RP11-466I1.1 |
| chr7_+_5465382 | 0.09 |
ENST00000609130.1
|
RP11-1275H24.2
|
RP11-1275H24.2 |
| chr6_+_31950150 | 0.09 |
ENST00000537134.1
|
C4A
|
complement component 4A (Rodgers blood group) |
| chr6_-_18155285 | 0.09 |
ENST00000309983.4
|
TPMT
|
thiopurine S-methyltransferase |
| chr3_+_140947563 | 0.09 |
ENST00000505013.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr12_-_6798410 | 0.09 |
ENST00000361959.3
ENST00000436774.2 ENST00000544482.1 |
ZNF384
|
zinc finger protein 384 |
| chr6_+_32938665 | 0.09 |
ENST00000374831.4
ENST00000395289.2 |
BRD2
|
bromodomain containing 2 |
| chr20_+_2673383 | 0.09 |
ENST00000380648.4
ENST00000342725.5 |
EBF4
|
early B-cell factor 4 |
| chr3_-_50605077 | 0.08 |
ENST00000426034.1
ENST00000441239.1 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr11_+_73676281 | 0.08 |
ENST00000543947.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr12_-_58240470 | 0.08 |
ENST00000548823.1
ENST00000398073.2 |
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr1_-_243417762 | 0.08 |
ENST00000522191.1
|
CEP170
|
centrosomal protein 170kDa |
| chr1_-_36023251 | 0.08 |
ENST00000426982.2
|
KIAA0319L
|
KIAA0319-like |
| chr12_+_56114151 | 0.08 |
ENST00000547072.1
ENST00000552930.1 ENST00000257895.5 |
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr11_+_125495862 | 0.08 |
ENST00000428830.2
ENST00000544373.1 ENST00000527013.1 ENST00000526937.1 ENST00000534685.1 |
CHEK1
|
checkpoint kinase 1 |
| chr1_-_53019059 | 0.08 |
ENST00000484723.2
ENST00000524582.1 |
ZCCHC11
|
zinc finger, CCHC domain containing 11 |
| chr14_-_51297837 | 0.08 |
ENST00000245441.5
ENST00000389868.3 ENST00000382041.3 ENST00000324330.9 ENST00000453196.1 ENST00000453401.2 |
NIN
|
ninein (GSK3B interacting protein) |
| chr1_+_29063119 | 0.08 |
ENST00000474884.1
ENST00000542507.1 |
YTHDF2
|
YTH domain family, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF263
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1905069 | allantois development(GO:1905069) |
| 0.1 | 0.2 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.1 | 0.5 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0060262 | regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.0 | 0.1 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.1 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.2 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.2 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.0 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.0 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.2 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.1 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.0 | 0.2 | GO:1904688 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.3 | GO:1902846 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.2 | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) |
| 0.0 | 0.2 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.0 | 0.0 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 | 0.1 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.1 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.1 | GO:0033363 | secretory granule organization(GO:0033363) |
| 0.0 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.3 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.0 | 0.1 | GO:0010160 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.3 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.1 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.2 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.2 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0002276 | basophil activation involved in immune response(GO:0002276) basophil activation(GO:0045575) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.2 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:1903762 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.0 | 0.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.1 | GO:0035261 | external genitalia morphogenesis(GO:0035261) |
| 0.0 | 0.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.1 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.1 | GO:0015965 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 | 0.1 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0035978 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.0 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.0 | GO:1902871 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.0 | GO:0038162 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.1 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.0 | 0.0 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.0 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.0 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0003064 | regulation of heart rate by hormone(GO:0003064) |
| 0.0 | 0.0 | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.0 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.1 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.0 | GO:1903280 | negative regulation of calcium:sodium antiporter activity(GO:1903280) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:1904478 | regulation of intestinal absorption(GO:1904478) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.0 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.0 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.0 | GO:0048372 | lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) |
| 0.0 | 0.0 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.4 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.0 | GO:0014011 | Schwann cell proliferation involved in axon regeneration(GO:0014011) negative regulation of Schwann cell migration(GO:1900148) regulation of Schwann cell proliferation involved in axon regeneration(GO:1905044) negative regulation of Schwann cell proliferation involved in axon regeneration(GO:1905045) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.0 | GO:1904784 | NLRP1 inflammasome complex assembly(GO:1904784) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0051414 | response to cortisol(GO:0051414) apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.0 | GO:0061054 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.0 | 0.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.2 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.3 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.0 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.3 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.2 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.1 | 0.2 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.1 | GO:0046921 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.0 | 0.1 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.2 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.0 | 0.1 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.1 | GO:0016855 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.1 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.3 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.1 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.0 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.0 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.0 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.0 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.0 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.0 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.0 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.3 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0035671 | enone reductase activity(GO:0035671) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 1.0 | GO:0004407 | histone deacetylase activity(GO:0004407) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |