Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
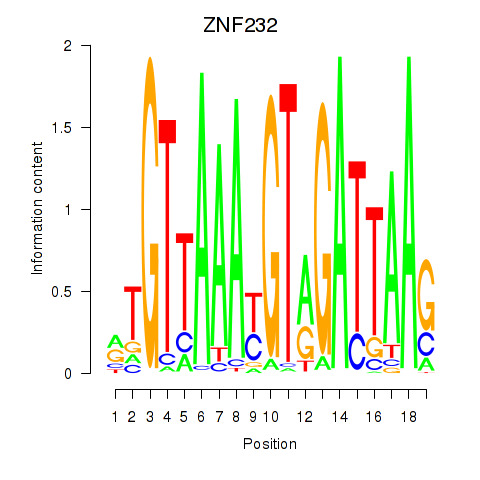
Results for ZNF232
Z-value: 0.45
Transcription factors associated with ZNF232
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF232
|
ENSG00000167840.9 | zinc finger protein 232 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF232 | hg19_v2_chr17_-_5026397_5026422 | 0.50 | 3.1e-01 | Click! |
Activity profile of ZNF232 motif
Sorted Z-values of ZNF232 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_153029980 | 0.48 |
ENST00000392653.2
|
SPRR2A
|
small proline-rich protein 2A |
| chr14_+_91709103 | 0.39 |
ENST00000553725.1
|
CTD-2547L24.3
|
HCG1816139; Uncharacterized protein |
| chr8_-_10336885 | 0.31 |
ENST00000520494.1
|
RP11-981G7.3
|
RP11-981G7.3 |
| chr16_+_30662085 | 0.22 |
ENST00000569864.1
|
PRR14
|
proline rich 14 |
| chr13_-_30160925 | 0.21 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr17_+_40458130 | 0.21 |
ENST00000587646.1
|
STAT5A
|
signal transducer and activator of transcription 5A |
| chr4_+_75174204 | 0.20 |
ENST00000332112.4
ENST00000514968.1 ENST00000503098.1 ENST00000502358.1 ENST00000509145.1 ENST00000505212.1 |
EPGN
|
epithelial mitogen |
| chr7_+_1609765 | 0.19 |
ENST00000437964.1
ENST00000533935.1 ENST00000532358.1 ENST00000524978.1 |
PSMG3-AS1
|
PSMG3 antisense RNA 1 (head to head) |
| chr19_+_18043810 | 0.18 |
ENST00000445755.2
|
CCDC124
|
coiled-coil domain containing 124 |
| chr2_-_31637560 | 0.17 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr2_+_234826016 | 0.15 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr19_-_14530112 | 0.12 |
ENST00000592632.1
ENST00000589675.1 |
DDX39A
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39A |
| chr20_+_4129426 | 0.12 |
ENST00000339123.6
ENST00000305958.4 ENST00000278795.3 |
SMOX
|
spermine oxidase |
| chr19_-_10491130 | 0.12 |
ENST00000530829.1
ENST00000529370.1 |
TYK2
|
tyrosine kinase 2 |
| chr4_-_84255935 | 0.12 |
ENST00000513463.1
|
HPSE
|
heparanase |
| chr1_-_193074504 | 0.12 |
ENST00000367439.3
|
GLRX2
|
glutaredoxin 2 |
| chr18_+_61637159 | 0.11 |
ENST00000397985.2
ENST00000353706.2 ENST00000542677.1 ENST00000397988.3 ENST00000448851.1 |
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| chr19_-_10491234 | 0.11 |
ENST00000524462.1
ENST00000531836.1 ENST00000525621.1 |
TYK2
|
tyrosine kinase 2 |
| chr16_+_30662050 | 0.10 |
ENST00000568754.1
|
PRR14
|
proline rich 14 |
| chr6_+_109416684 | 0.10 |
ENST00000521522.1
ENST00000524064.1 ENST00000522608.1 ENST00000521503.1 ENST00000519407.1 ENST00000519095.1 ENST00000368968.2 ENST00000522490.1 ENST00000523209.1 ENST00000368970.2 ENST00000520883.1 ENST00000523787.1 |
CEP57L1
|
centrosomal protein 57kDa-like 1 |
| chr2_-_225811747 | 0.08 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr3_-_130745403 | 0.08 |
ENST00000504725.1
ENST00000509060.1 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr16_+_30662360 | 0.08 |
ENST00000542965.2
|
PRR14
|
proline rich 14 |
| chr14_-_24711865 | 0.07 |
ENST00000399423.4
ENST00000267415.7 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr22_-_30234218 | 0.07 |
ENST00000307790.3
ENST00000542393.1 ENST00000397771.2 |
ASCC2
|
activating signal cointegrator 1 complex subunit 2 |
| chr16_+_30662184 | 0.07 |
ENST00000300835.4
|
PRR14
|
proline rich 14 |
| chr9_+_5231413 | 0.07 |
ENST00000239316.4
|
INSL4
|
insulin-like 4 (placenta) |
| chr7_-_1609591 | 0.07 |
ENST00000288607.2
ENST00000404674.3 |
PSMG3
|
proteasome (prosome, macropain) assembly chaperone 3 |
| chr16_-_56223480 | 0.07 |
ENST00000565155.1
|
RP11-461O7.1
|
RP11-461O7.1 |
| chr18_+_55816546 | 0.07 |
ENST00000435432.2
ENST00000357895.5 ENST00000586263.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr2_+_74425689 | 0.06 |
ENST00000394053.2
ENST00000409804.1 ENST00000264090.4 ENST00000394050.3 ENST00000409601.1 |
MTHFD2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr7_+_142829162 | 0.06 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr14_-_21516590 | 0.06 |
ENST00000555026.1
|
NDRG2
|
NDRG family member 2 |
| chr14_-_24711806 | 0.06 |
ENST00000540705.1
ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr3_+_8543393 | 0.06 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr16_-_4852915 | 0.05 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr2_-_74619152 | 0.05 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr15_-_74374891 | 0.05 |
ENST00000290438.3
|
GOLGA6A
|
golgin A6 family, member A |
| chr14_-_24711764 | 0.05 |
ENST00000557921.1
ENST00000558476.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr1_+_84944926 | 0.05 |
ENST00000370656.1
ENST00000370654.5 |
RPF1
|
ribosome production factor 1 homolog (S. cerevisiae) |
| chr8_+_22601 | 0.05 |
ENST00000522481.3
ENST00000518652.1 |
AC144568.2
|
Uncharacterized protein |
| chr3_+_8543533 | 0.05 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr6_+_109416313 | 0.05 |
ENST00000521277.1
ENST00000517392.1 ENST00000407272.1 ENST00000336977.4 ENST00000519286.1 ENST00000518329.1 ENST00000522461.1 ENST00000518853.1 |
CEP57L1
|
centrosomal protein 57kDa-like 1 |
| chr6_-_26056695 | 0.05 |
ENST00000343677.2
|
HIST1H1C
|
histone cluster 1, H1c |
| chr11_-_117800080 | 0.05 |
ENST00000524993.1
ENST00000528626.1 ENST00000445164.2 ENST00000430170.2 ENST00000526090.1 |
TMPRSS13
|
transmembrane protease, serine 13 |
| chr3_+_184018352 | 0.04 |
ENST00000435761.1
ENST00000439383.1 |
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chrY_+_15016725 | 0.04 |
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr2_+_228735763 | 0.04 |
ENST00000373666.2
|
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr19_-_50380536 | 0.04 |
ENST00000391832.3
ENST00000391834.2 ENST00000344175.5 |
AKT1S1
|
AKT1 substrate 1 (proline-rich) |
| chr1_+_156105878 | 0.03 |
ENST00000508500.1
|
LMNA
|
lamin A/C |
| chr1_-_173572181 | 0.03 |
ENST00000536496.1
ENST00000367714.3 |
SLC9C2
|
solute carrier family 9, member C2 (putative) |
| chr15_-_34635314 | 0.03 |
ENST00000557912.1
ENST00000328848.4 |
NOP10
|
NOP10 ribonucleoprotein |
| chrX_+_100805496 | 0.03 |
ENST00000372829.3
|
ARMCX1
|
armadillo repeat containing, X-linked 1 |
| chrY_+_15016013 | 0.03 |
ENST00000360160.4
ENST00000454054.1 |
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr2_+_102413726 | 0.03 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr17_-_3301704 | 0.03 |
ENST00000322608.2
|
OR1E1
|
olfactory receptor, family 1, subfamily E, member 1 |
| chr17_+_39975544 | 0.02 |
ENST00000544340.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr17_-_6915646 | 0.02 |
ENST00000574377.1
ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2
|
Uncharacterized protein |
| chr2_-_74618964 | 0.02 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr1_-_36107445 | 0.02 |
ENST00000373237.3
|
PSMB2
|
proteasome (prosome, macropain) subunit, beta type, 2 |
| chr1_+_237205476 | 0.02 |
ENST00000366574.2
|
RYR2
|
ryanodine receptor 2 (cardiac) |
| chr9_-_107367951 | 0.02 |
ENST00000542196.1
|
OR13C2
|
olfactory receptor, family 13, subfamily C, member 2 |
| chrX_+_36065053 | 0.02 |
ENST00000313548.4
|
CHDC2
|
calponin homology domain containing 2 |
| chrX_+_2924654 | 0.02 |
ENST00000381130.2
|
ARSH
|
arylsulfatase family, member H |
| chr17_+_39975455 | 0.02 |
ENST00000455106.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr5_+_177435986 | 0.01 |
ENST00000398106.2
|
FAM153C
|
family with sequence similarity 153, member C |
| chr4_-_88450612 | 0.01 |
ENST00000418378.1
ENST00000282470.6 |
SPARCL1
|
SPARC-like 1 (hevin) |
| chr11_+_66276550 | 0.01 |
ENST00000419755.3
|
CTD-3074O7.11
|
Bardet-Biedl syndrome 1 protein |
| chr1_-_79472365 | 0.01 |
ENST00000370742.3
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1 |
| chr17_+_26800296 | 0.01 |
ENST00000444914.3
ENST00000314669.5 |
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr10_+_134150835 | 0.01 |
ENST00000432555.2
|
LRRC27
|
leucine rich repeat containing 27 |
| chr9_+_117085336 | 0.00 |
ENST00000259396.8
ENST00000538816.1 |
ORM1
|
orosomucoid 1 |
| chr10_-_115904361 | 0.00 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr17_+_26800648 | 0.00 |
ENST00000545060.1
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr16_-_11367452 | 0.00 |
ENST00000327157.2
|
PRM3
|
protamine 3 |
| chr1_-_204654826 | 0.00 |
ENST00000367177.3
|
LRRN2
|
leucine rich repeat neuronal 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF232
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.2 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.2 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.2 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.0 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.0 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.0 | GO:0034515 | proteasome storage granule(GO:0034515) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0052894 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.2 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |