Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
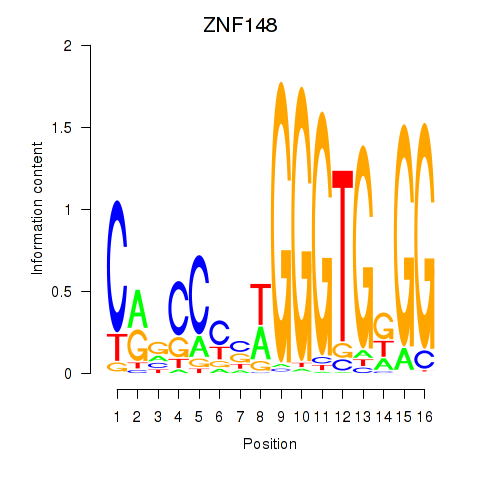
Results for ZNF148
Z-value: 0.95
Transcription factors associated with ZNF148
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF148
|
ENSG00000163848.14 | zinc finger protein 148 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF148 | hg19_v2_chr3_-_125094093_125094198 | -0.79 | 6.1e-02 | Click! |
Activity profile of ZNF148 motif
Sorted Z-values of ZNF148 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_43968306 | 0.86 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr19_-_42192189 | 0.65 |
ENST00000401731.1
ENST00000338196.4 ENST00000006724.3 |
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr8_+_104383759 | 0.58 |
ENST00000415886.2
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr15_-_76352069 | 0.55 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr20_+_62185491 | 0.51 |
ENST00000370097.1
|
C20orf195
|
chromosome 20 open reading frame 195 |
| chr10_+_120967072 | 0.45 |
ENST00000392870.2
|
GRK5
|
G protein-coupled receptor kinase 5 |
| chr19_+_54371114 | 0.44 |
ENST00000448420.1
ENST00000439000.1 ENST00000391770.4 ENST00000391771.1 |
MYADM
|
myeloid-associated differentiation marker |
| chr10_-_75532373 | 0.42 |
ENST00000595757.1
|
AC022400.2
|
Uncharacterized protein; cDNA FLJ44715 fis, clone BRACE3021430 |
| chr11_+_46402482 | 0.38 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr3_-_49203744 | 0.38 |
ENST00000321895.6
|
CCDC71
|
coiled-coil domain containing 71 |
| chr19_-_663171 | 0.36 |
ENST00000606896.1
ENST00000589762.2 |
RNF126
|
ring finger protein 126 |
| chr17_-_27188984 | 0.36 |
ENST00000582320.2
|
MIR144
|
microRNA 451b |
| chr6_-_41908428 | 0.35 |
ENST00000505064.1
|
CCND3
|
cyclin D3 |
| chr22_+_38382163 | 0.35 |
ENST00000333418.4
ENST00000427034.1 |
POLR2F
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr1_-_63782888 | 0.34 |
ENST00000436475.2
|
LINC00466
|
long intergenic non-protein coding RNA 466 |
| chr8_+_104383728 | 0.34 |
ENST00000330295.5
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr2_-_27333965 | 0.33 |
ENST00000452318.2
|
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr20_+_19738792 | 0.32 |
ENST00000412571.1
|
RP1-122P22.2
|
RP1-122P22.2 |
| chr19_-_42916499 | 0.32 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr19_+_54369434 | 0.31 |
ENST00000421337.1
|
MYADM
|
myeloid-associated differentiation marker |
| chr5_+_172484377 | 0.31 |
ENST00000523161.1
|
CREBRF
|
CREB3 regulatory factor |
| chr19_-_12267524 | 0.31 |
ENST00000455799.1
ENST00000355738.1 ENST00000439556.2 ENST00000542938.1 |
ZNF625
|
zinc finger protein 625 |
| chr20_-_36793663 | 0.30 |
ENST00000536701.1
ENST00000536724.1 |
TGM2
|
transglutaminase 2 |
| chr1_+_161068179 | 0.30 |
ENST00000368011.4
ENST00000392192.2 |
KLHDC9
|
kelch domain containing 9 |
| chr8_-_11873043 | 0.30 |
ENST00000527396.1
|
RP11-481A20.11
|
Protein LOC101060662 |
| chr14_+_94492674 | 0.29 |
ENST00000203664.5
ENST00000553723.1 |
OTUB2
|
OTU domain, ubiquitin aldehyde binding 2 |
| chr5_+_71403280 | 0.29 |
ENST00000511641.2
|
MAP1B
|
microtubule-associated protein 1B |
| chr8_-_91618285 | 0.29 |
ENST00000517505.1
|
LINC01030
|
long intergenic non-protein coding RNA 1030 |
| chr1_+_44444865 | 0.29 |
ENST00000372324.1
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr12_+_120933859 | 0.28 |
ENST00000242577.6
ENST00000548214.1 ENST00000392508.2 |
DYNLL1
|
dynein, light chain, LC8-type 1 |
| chr1_+_44445643 | 0.27 |
ENST00000309519.7
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr20_-_36793774 | 0.27 |
ENST00000361475.2
|
TGM2
|
transglutaminase 2 |
| chr19_-_18717627 | 0.27 |
ENST00000392386.3
|
CRLF1
|
cytokine receptor-like factor 1 |
| chr6_-_38607628 | 0.26 |
ENST00000498633.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr6_+_144471643 | 0.25 |
ENST00000367568.4
|
STX11
|
syntaxin 11 |
| chr19_-_45579762 | 0.25 |
ENST00000303809.2
|
ZNF296
|
zinc finger protein 296 |
| chr19_+_797392 | 0.24 |
ENST00000350092.4
ENST00000349038.4 ENST00000586481.1 ENST00000585535.1 |
PTBP1
|
polypyrimidine tract binding protein 1 |
| chr4_+_6271558 | 0.24 |
ENST00000503569.1
ENST00000226760.1 |
WFS1
|
Wolfram syndrome 1 (wolframin) |
| chr14_-_88200641 | 0.24 |
ENST00000556168.1
|
RP11-1152H15.1
|
RP11-1152H15.1 |
| chr12_-_49581152 | 0.24 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr19_+_44037546 | 0.23 |
ENST00000601282.1
|
ZNF575
|
zinc finger protein 575 |
| chr22_-_20138302 | 0.23 |
ENST00000540078.1
ENST00000439765.2 |
AC006547.14
|
uncharacterized protein LOC388849 |
| chr22_-_50913371 | 0.23 |
ENST00000348911.6
ENST00000380817.3 ENST00000390679.3 |
SBF1
|
SET binding factor 1 |
| chr22_-_20792089 | 0.23 |
ENST00000405555.3
ENST00000266214.5 |
SCARF2
|
scavenger receptor class F, member 2 |
| chr15_+_65134088 | 0.23 |
ENST00000323544.4
ENST00000437723.1 |
PLEKHO2
AC069368.3
|
pleckstrin homology domain containing, family O member 2 Uncharacterized protein |
| chr9_+_140149625 | 0.23 |
ENST00000343053.4
|
NELFB
|
negative elongation factor complex member B |
| chr17_+_6918354 | 0.23 |
ENST00000552775.1
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr5_-_176738883 | 0.22 |
ENST00000513169.1
ENST00000423571.2 ENST00000502529.1 ENST00000427908.2 |
MXD3
|
MAX dimerization protein 3 |
| chr2_+_37571717 | 0.22 |
ENST00000338415.3
ENST00000404976.1 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr1_-_89738528 | 0.22 |
ENST00000343435.5
|
GBP5
|
guanylate binding protein 5 |
| chr15_+_31619013 | 0.22 |
ENST00000307145.3
|
KLF13
|
Kruppel-like factor 13 |
| chr5_+_71403061 | 0.22 |
ENST00000512974.1
ENST00000296755.7 |
MAP1B
|
microtubule-associated protein 1B |
| chr9_+_6716478 | 0.21 |
ENST00000452643.1
|
RP11-390F4.3
|
RP11-390F4.3 |
| chr19_-_14247365 | 0.21 |
ENST00000592798.1
ENST00000474890.1 |
ASF1B
|
anti-silencing function 1B histone chaperone |
| chr12_-_49582593 | 0.21 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr12_+_57998400 | 0.21 |
ENST00000548804.1
ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr1_+_156698743 | 0.21 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr10_+_134000404 | 0.20 |
ENST00000338492.4
ENST00000368629.1 |
DPYSL4
|
dihydropyrimidinase-like 4 |
| chr16_+_50313426 | 0.20 |
ENST00000569265.1
|
ADCY7
|
adenylate cyclase 7 |
| chrX_-_39923656 | 0.20 |
ENST00000413905.1
|
BCOR
|
BCL6 corepressor |
| chr9_-_35732362 | 0.20 |
ENST00000314888.9
ENST00000540444.1 |
TLN1
|
talin 1 |
| chr19_-_6591113 | 0.20 |
ENST00000423145.3
ENST00000245903.3 |
CD70
|
CD70 molecule |
| chr12_-_49582978 | 0.20 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr12_+_132312931 | 0.19 |
ENST00000360564.1
ENST00000545671.1 ENST00000545790.1 |
MMP17
|
matrix metallopeptidase 17 (membrane-inserted) |
| chr16_+_2732476 | 0.19 |
ENST00000301738.4
ENST00000564195.1 |
KCTD5
|
potassium channel tetramerization domain containing 5 |
| chr8_-_67525524 | 0.19 |
ENST00000517885.1
|
MYBL1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr17_-_6616678 | 0.19 |
ENST00000381074.4
ENST00000293800.6 ENST00000572352.1 ENST00000576323.1 ENST00000573648.1 |
SLC13A5
|
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr19_+_7985880 | 0.19 |
ENST00000597584.1
|
SNAPC2
|
small nuclear RNA activating complex, polypeptide 2, 45kDa |
| chr11_+_76493294 | 0.19 |
ENST00000533752.1
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr3_-_39196049 | 0.19 |
ENST00000514182.1
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1 |
| chr9_+_74764278 | 0.19 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chr22_-_38669030 | 0.19 |
ENST00000361906.3
|
TMEM184B
|
transmembrane protein 184B |
| chr5_-_176900610 | 0.18 |
ENST00000477391.2
ENST00000393565.1 ENST00000309007.5 |
DBN1
|
drebrin 1 |
| chr16_+_58035277 | 0.18 |
ENST00000219281.3
ENST00000539737.2 ENST00000423271.3 ENST00000561568.1 ENST00000563149.1 |
USB1
|
U6 snRNA biogenesis 1 |
| chr1_+_35225339 | 0.18 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr3_-_88108212 | 0.18 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr12_-_56693758 | 0.18 |
ENST00000547298.1
ENST00000551936.1 ENST00000551253.1 ENST00000551473.1 |
CS
|
citrate synthase |
| chr6_-_31763276 | 0.18 |
ENST00000440048.1
|
VARS
|
valyl-tRNA synthetase |
| chr3_+_57994127 | 0.18 |
ENST00000490882.1
ENST00000295956.4 ENST00000358537.3 ENST00000429972.2 ENST00000348383.5 ENST00000357272.4 |
FLNB
|
filamin B, beta |
| chr12_-_49582837 | 0.18 |
ENST00000547939.1
ENST00000546918.1 ENST00000552924.1 |
TUBA1A
|
tubulin, alpha 1a |
| chr18_+_55102917 | 0.18 |
ENST00000491143.2
|
ONECUT2
|
one cut homeobox 2 |
| chr2_+_239335636 | 0.17 |
ENST00000409297.1
|
ASB1
|
ankyrin repeat and SOCS box containing 1 |
| chr15_-_74494779 | 0.17 |
ENST00000571341.1
|
STRA6
|
stimulated by retinoic acid 6 |
| chr6_+_83903061 | 0.17 |
ENST00000369724.4
ENST00000539997.1 |
RWDD2A
|
RWD domain containing 2A |
| chr6_+_31633833 | 0.17 |
ENST00000375882.2
ENST00000375880.2 |
CSNK2B
CSNK2B-LY6G5B-1181
|
casein kinase 2, beta polypeptide Uncharacterized protein |
| chr1_-_1208851 | 0.17 |
ENST00000488418.1
|
UBE2J2
|
ubiquitin-conjugating enzyme E2, J2 |
| chr20_-_3748416 | 0.16 |
ENST00000399672.1
|
C20orf27
|
chromosome 20 open reading frame 27 |
| chr6_+_35310312 | 0.16 |
ENST00000448077.2
ENST00000360694.3 ENST00000418635.2 ENST00000444397.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr9_+_116638562 | 0.16 |
ENST00000374126.5
ENST00000288466.7 |
ZNF618
|
zinc finger protein 618 |
| chr20_-_3748363 | 0.16 |
ENST00000217195.8
|
C20orf27
|
chromosome 20 open reading frame 27 |
| chr2_+_181845532 | 0.16 |
ENST00000602475.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr22_+_39795746 | 0.16 |
ENST00000216160.6
ENST00000331454.3 |
TAB1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr9_+_35732312 | 0.16 |
ENST00000353704.2
|
CREB3
|
cAMP responsive element binding protein 3 |
| chr11_+_12132117 | 0.15 |
ENST00000256194.4
|
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr2_-_39187479 | 0.15 |
ENST00000601251.1
|
AC019171.1
|
Uncharacterized protein |
| chr19_-_19626838 | 0.15 |
ENST00000360913.3
|
TSSK6
|
testis-specific serine kinase 6 |
| chr2_+_85132749 | 0.15 |
ENST00000233143.4
|
TMSB10
|
thymosin beta 10 |
| chr12_+_57998595 | 0.15 |
ENST00000337737.3
ENST00000548198.1 ENST00000551632.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr22_+_35653445 | 0.15 |
ENST00000420166.1
ENST00000444518.2 ENST00000455359.1 ENST00000216106.5 |
HMGXB4
|
HMG box domain containing 4 |
| chr16_-_58034357 | 0.15 |
ENST00000562909.1
|
ZNF319
|
zinc finger protein 319 |
| chr6_-_44095183 | 0.15 |
ENST00000372014.3
|
MRPL14
|
mitochondrial ribosomal protein L14 |
| chr16_+_89894875 | 0.15 |
ENST00000393062.2
|
SPIRE2
|
spire-type actin nucleation factor 2 |
| chr6_-_127840453 | 0.15 |
ENST00000556132.1
|
SOGA3
|
SOGA family member 3 |
| chr1_+_33546714 | 0.15 |
ENST00000294517.6
ENST00000358680.3 ENST00000373443.3 ENST00000398167.1 |
ADC
|
arginine decarboxylase |
| chr19_-_49243845 | 0.15 |
ENST00000222145.4
|
RASIP1
|
Ras interacting protein 1 |
| chr10_-_30348439 | 0.15 |
ENST00000375377.1
|
KIAA1462
|
KIAA1462 |
| chrX_+_47078069 | 0.15 |
ENST00000357227.4
ENST00000519758.1 ENST00000520893.1 ENST00000517426.1 |
CDK16
|
cyclin-dependent kinase 16 |
| chr20_-_44539538 | 0.15 |
ENST00000372420.1
|
PLTP
|
phospholipid transfer protein |
| chr12_-_90103077 | 0.15 |
ENST00000551310.1
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr19_-_55740632 | 0.15 |
ENST00000327042.4
|
TMEM86B
|
transmembrane protein 86B |
| chr12_-_56694083 | 0.14 |
ENST00000552688.1
ENST00000548041.1 ENST00000551137.1 ENST00000551968.1 ENST00000542324.2 ENST00000546930.1 ENST00000549221.1 ENST00000550159.1 ENST00000550734.1 |
CS
|
citrate synthase |
| chr20_+_61904199 | 0.14 |
ENST00000518601.2
ENST00000353546.3 |
ARFGAP1
|
ADP-ribosylation factor GTPase activating protein 1 |
| chr18_+_43684310 | 0.14 |
ENST00000592471.1
ENST00000585518.1 |
HAUS1
|
HAUS augmin-like complex, subunit 1 |
| chr11_+_119038897 | 0.14 |
ENST00000454811.1
ENST00000449394.1 |
NLRX1
|
NLR family member X1 |
| chr21_-_46330545 | 0.14 |
ENST00000320216.6
ENST00000397852.1 |
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr6_+_35310391 | 0.14 |
ENST00000337400.2
ENST00000311565.4 ENST00000540939.1 |
PPARD
|
peroxisome proliferator-activated receptor delta |
| chr1_-_43833628 | 0.14 |
ENST00000413844.2
ENST00000372458.3 |
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr19_+_797443 | 0.14 |
ENST00000394601.4
ENST00000589575.1 |
PTBP1
|
polypyrimidine tract binding protein 1 |
| chr3_+_184032919 | 0.14 |
ENST00000427845.1
ENST00000342981.4 ENST00000319274.6 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr17_-_34890759 | 0.14 |
ENST00000431794.3
|
MYO19
|
myosin XIX |
| chr12_+_120933904 | 0.14 |
ENST00000550178.1
ENST00000550845.1 ENST00000549989.1 ENST00000552870.1 |
DYNLL1
|
dynein, light chain, LC8-type 1 |
| chr17_+_1945301 | 0.14 |
ENST00000572195.1
|
OVCA2
|
ovarian tumor suppressor candidate 2 |
| chr2_-_241831424 | 0.13 |
ENST00000402775.2
ENST00000307486.8 |
C2orf54
|
chromosome 2 open reading frame 54 |
| chr1_-_8483723 | 0.13 |
ENST00000476556.1
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr7_+_100273736 | 0.13 |
ENST00000412215.1
ENST00000393924.1 |
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr4_-_105416039 | 0.13 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr12_+_6898674 | 0.13 |
ENST00000541982.1
ENST00000539492.1 |
CD4
|
CD4 molecule |
| chr6_-_31651817 | 0.13 |
ENST00000375863.3
ENST00000375860.2 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr17_+_17206635 | 0.13 |
ENST00000389022.4
|
NT5M
|
5',3'-nucleotidase, mitochondrial |
| chr15_+_66994663 | 0.13 |
ENST00000457357.2
|
SMAD6
|
SMAD family member 6 |
| chr9_-_34665983 | 0.13 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr14_+_105331596 | 0.13 |
ENST00000556508.1
ENST00000414716.3 ENST00000453495.1 ENST00000418279.1 |
CEP170B
|
centrosomal protein 170B |
| chr2_-_71221942 | 0.13 |
ENST00000272438.4
|
TEX261
|
testis expressed 261 |
| chr9_+_130565147 | 0.13 |
ENST00000373247.2
ENST00000373245.1 ENST00000393706.2 ENST00000373228.1 |
FPGS
|
folylpolyglutamate synthase |
| chr1_+_2985760 | 0.13 |
ENST00000378391.2
ENST00000514189.1 ENST00000270722.5 |
PRDM16
|
PR domain containing 16 |
| chr9_+_116638630 | 0.13 |
ENST00000452710.1
ENST00000374124.4 |
ZNF618
|
zinc finger protein 618 |
| chr5_-_176037105 | 0.13 |
ENST00000303991.4
|
GPRIN1
|
G protein regulated inducer of neurite outgrowth 1 |
| chr16_-_776431 | 0.13 |
ENST00000293889.6
|
CCDC78
|
coiled-coil domain containing 78 |
| chr8_+_32406179 | 0.13 |
ENST00000405005.3
|
NRG1
|
neuregulin 1 |
| chr16_-_24942655 | 0.13 |
ENST00000573765.1
|
ARHGAP17
|
Rho GTPase activating protein 17 |
| chr14_+_31343951 | 0.13 |
ENST00000556908.1
ENST00000555881.1 ENST00000460581.2 |
COCH
|
cochlin |
| chr11_-_34938039 | 0.13 |
ENST00000395787.3
|
APIP
|
APAF1 interacting protein |
| chr22_+_45098067 | 0.13 |
ENST00000336985.6
ENST00000403696.1 ENST00000457960.1 ENST00000361473.5 |
PRR5
PRR5-ARHGAP8
|
proline rich 5 (renal) PRR5-ARHGAP8 readthrough |
| chr17_-_7197881 | 0.13 |
ENST00000007699.5
|
YBX2
|
Y box binding protein 2 |
| chr20_+_61904137 | 0.13 |
ENST00000370283.4
ENST00000523114.1 ENST00000547204.1 ENST00000549047.1 ENST00000523460.1 ENST00000519604.1 ENST00000519273.2 ENST00000370275.4 |
ARFGAP1
|
ADP-ribosylation factor GTPase activating protein 1 |
| chr19_-_42192096 | 0.12 |
ENST00000602225.1
|
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr8_-_135725205 | 0.12 |
ENST00000523399.1
ENST00000377838.3 |
ZFAT
|
zinc finger and AT hook domain containing |
| chr14_+_24105544 | 0.12 |
ENST00000250383.6
ENST00000344777.7 |
DHRS2
|
dehydrogenase/reductase (SDR family) member 2 |
| chr3_-_195538760 | 0.12 |
ENST00000475231.1
|
MUC4
|
mucin 4, cell surface associated |
| chr5_+_52776228 | 0.12 |
ENST00000256759.3
|
FST
|
follistatin |
| chr5_+_141016508 | 0.12 |
ENST00000444782.1
ENST00000521367.1 ENST00000297164.3 |
RELL2
|
RELT-like 2 |
| chr3_-_52567792 | 0.12 |
ENST00000307092.4
ENST00000422318.2 ENST00000459839.1 |
NT5DC2
|
5'-nucleotidase domain containing 2 |
| chr9_-_35618364 | 0.12 |
ENST00000378431.1
ENST00000378430.3 ENST00000259633.4 |
CD72
|
CD72 molecule |
| chr6_+_15401075 | 0.12 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr17_-_1394940 | 0.12 |
ENST00000570984.2
ENST00000361007.2 |
MYO1C
|
myosin IC |
| chr11_+_64009072 | 0.12 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr14_+_77843459 | 0.12 |
ENST00000216471.4
|
SAMD15
|
sterile alpha motif domain containing 15 |
| chr1_-_95007193 | 0.12 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr19_-_13044494 | 0.12 |
ENST00000593021.1
ENST00000587981.1 ENST00000423140.2 ENST00000314606.4 |
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr1_+_155829341 | 0.12 |
ENST00000539162.1
|
SYT11
|
synaptotagmin XI |
| chrX_-_152989798 | 0.12 |
ENST00000441714.1
ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31
|
B-cell receptor-associated protein 31 |
| chr1_+_167190066 | 0.12 |
ENST00000367866.2
ENST00000429375.2 ENST00000452019.1 ENST00000420254.3 ENST00000541643.3 |
POU2F1
|
POU class 2 homeobox 1 |
| chr3_+_108541545 | 0.12 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr16_-_28074822 | 0.12 |
ENST00000395724.3
ENST00000380898.2 ENST00000447459.2 |
GSG1L
|
GSG1-like |
| chr16_+_8806800 | 0.12 |
ENST00000561870.1
ENST00000396600.2 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr16_-_4838255 | 0.12 |
ENST00000591624.1
ENST00000396693.5 |
SEPT12
|
septin 12 |
| chr16_+_29984962 | 0.12 |
ENST00000308893.4
|
TAOK2
|
TAO kinase 2 |
| chr2_-_100721178 | 0.12 |
ENST00000409236.2
|
AFF3
|
AF4/FMR2 family, member 3 |
| chr3_-_49941042 | 0.12 |
ENST00000344206.4
ENST00000296474.3 |
MST1R
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
| chr11_+_46402583 | 0.12 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr19_-_40324767 | 0.12 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr12_-_120632505 | 0.11 |
ENST00000300648.6
|
GCN1L1
|
GCN1 general control of amino-acid synthesis 1-like 1 (yeast) |
| chr7_+_128470431 | 0.11 |
ENST00000325888.8
ENST00000346177.6 |
FLNC
|
filamin C, gamma |
| chr2_+_64681103 | 0.11 |
ENST00000464281.1
|
LGALSL
|
lectin, galactoside-binding-like |
| chr6_-_32151999 | 0.11 |
ENST00000375069.3
ENST00000538695.1 ENST00000438221.2 ENST00000375065.5 ENST00000450110.1 ENST00000375067.3 ENST00000375056.2 |
AGER
|
advanced glycosylation end product-specific receptor |
| chr8_-_123793048 | 0.11 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr15_-_40545111 | 0.11 |
ENST00000319503.3
ENST00000559727.1 |
C15orf56
|
chromosome 15 open reading frame 56 |
| chr6_-_127840336 | 0.11 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr3_+_184033135 | 0.11 |
ENST00000424196.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr9_-_130661916 | 0.11 |
ENST00000373142.1
ENST00000373146.1 ENST00000373144.3 |
ST6GALNAC6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr12_+_56522001 | 0.11 |
ENST00000267113.4
ENST00000541590.1 |
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr5_+_154135029 | 0.11 |
ENST00000518297.1
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr1_-_156698591 | 0.11 |
ENST00000368219.1
|
ISG20L2
|
interferon stimulated exonuclease gene 20kDa-like 2 |
| chr14_+_31343747 | 0.11 |
ENST00000216361.4
ENST00000396618.3 ENST00000475087.1 |
COCH
|
cochlin |
| chr19_+_50180507 | 0.11 |
ENST00000454376.2
ENST00000524771.1 |
PRMT1
|
protein arginine methyltransferase 1 |
| chr1_+_27022485 | 0.11 |
ENST00000324856.7
|
ARID1A
|
AT rich interactive domain 1A (SWI-like) |
| chr1_-_155224699 | 0.11 |
ENST00000491082.1
|
FAM189B
|
family with sequence similarity 189, member B |
| chr1_-_53686261 | 0.11 |
ENST00000294360.4
|
C1orf123
|
chromosome 1 open reading frame 123 |
| chr17_-_2615031 | 0.11 |
ENST00000576885.1
ENST00000574426.2 |
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr19_+_7599597 | 0.11 |
ENST00000414982.3
ENST00000450331.3 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr4_-_84255935 | 0.11 |
ENST00000513463.1
|
HPSE
|
heparanase |
| chr5_-_180237082 | 0.11 |
ENST00000506889.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr16_+_4838393 | 0.11 |
ENST00000589721.1
|
SMIM22
|
small integral membrane protein 22 |
| chr19_+_54960790 | 0.11 |
ENST00000443957.1
|
LENG8
|
leukocyte receptor cluster (LRC) member 8 |
| chr3_-_48130314 | 0.11 |
ENST00000439356.1
ENST00000395734.3 ENST00000426837.2 |
MAP4
|
microtubule-associated protein 4 |
| chr12_-_10766184 | 0.10 |
ENST00000539554.1
ENST00000381881.2 ENST00000320756.2 |
MAGOHB
|
mago-nashi homolog B (Drosophila) |
| chr2_+_239335449 | 0.10 |
ENST00000264607.4
|
ASB1
|
ankyrin repeat and SOCS box containing 1 |
| chr19_-_11688951 | 0.10 |
ENST00000589792.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr14_+_105886150 | 0.10 |
ENST00000331320.7
ENST00000406191.1 |
MTA1
|
metastasis associated 1 |
| chr20_-_31124186 | 0.10 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF148
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.1 | 0.6 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 | 0.9 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 0.8 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.3 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 0.6 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.3 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.1 | 0.1 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.1 | 0.2 | GO:0072616 | interleukin-18 secretion(GO:0072616) |
| 0.1 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.2 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.1 | 0.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.2 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.1 | 0.5 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.1 | 0.3 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.4 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.3 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.2 | GO:0035397 | helper T cell enhancement of adaptive immune response(GO:0035397) |
| 0.0 | 0.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:1904604 | regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904596) negative regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904597) regulation of advanced glycation end-product receptor activity(GO:1904603) negative regulation of advanced glycation end-product receptor activity(GO:1904604) negative regulation of connective tissue replacement(GO:1905204) |
| 0.0 | 0.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.5 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.2 | GO:1904450 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.2 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.0 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.2 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.2 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.4 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.1 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.5 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.1 | GO:0050822 | peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.0 | 0.2 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.1 | GO:0035283 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.3 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.3 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.2 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.2 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.1 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 0.8 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.2 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 0.1 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.5 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.1 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.2 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.2 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0046078 | dUMP metabolic process(GO:0046078) |
| 0.0 | 0.0 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.1 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.3 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:1900159 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) regulation of bone mineralization involved in bone maturation(GO:1900157) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.0 | 0.0 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.0 | 0.0 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.3 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.0 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.1 | GO:0072104 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.2 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.2 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.0 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.4 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.4 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.1 | 0.3 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.3 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.3 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.1 | 0.2 | GO:0070698 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) type I activin receptor binding(GO:0070698) |
| 0.1 | 0.2 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.1 | 0.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:1904599 | advanced glycation end-product binding(GO:1904599) |
| 0.0 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.2 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.1 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0017153 | citrate transmembrane transporter activity(GO:0015137) succinate transmembrane transporter activity(GO:0015141) tricarboxylic acid transmembrane transporter activity(GO:0015142) sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.1 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.3 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.3 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.0 | 0.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.2 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.3 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.0 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.4 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.1 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.0 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0051538 | iron-responsive element binding(GO:0030350) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.3 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.7 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |