Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
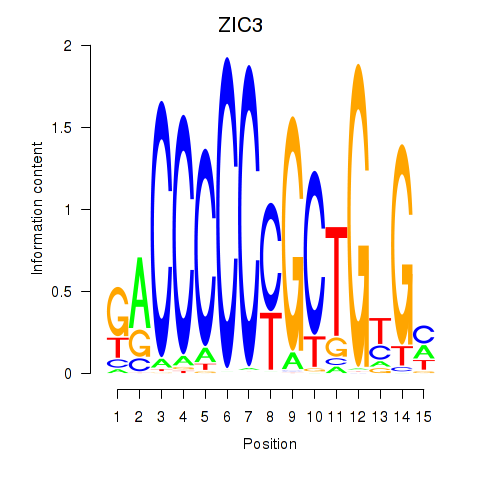
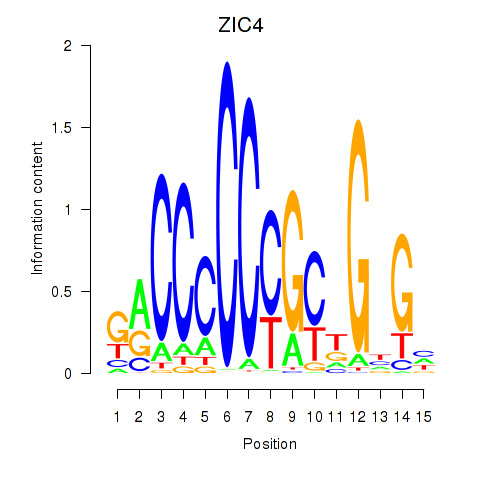
Results for ZIC3_ZIC4
Z-value: 0.76
Transcription factors associated with ZIC3_ZIC4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZIC3
|
ENSG00000156925.7 | Zic family member 3 |
|
ZIC4
|
ENSG00000174963.13 | Zic family member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZIC3 | hg19_v2_chrX_+_136648643_136648711 | 0.46 | 3.6e-01 | Click! |
Activity profile of ZIC3_ZIC4 motif
Sorted Z-values of ZIC3_ZIC4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_132312931 | 0.57 |
ENST00000360564.1
ENST00000545671.1 ENST00000545790.1 |
MMP17
|
matrix metallopeptidase 17 (membrane-inserted) |
| chr9_-_86955598 | 0.50 |
ENST00000376238.4
|
SLC28A3
|
solute carrier family 28 (concentrative nucleoside transporter), member 3 |
| chr9_+_19408919 | 0.44 |
ENST00000380376.1
|
ACER2
|
alkaline ceramidase 2 |
| chr19_+_45251804 | 0.41 |
ENST00000164227.5
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr11_-_67272794 | 0.37 |
ENST00000436757.2
ENST00000356404.3 |
PITPNM1
|
phosphatidylinositol transfer protein, membrane-associated 1 |
| chr20_+_34203794 | 0.36 |
ENST00000374273.3
|
SPAG4
|
sperm associated antigen 4 |
| chr7_-_994302 | 0.36 |
ENST00000265846.5
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr5_+_76506706 | 0.34 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr11_+_69061594 | 0.30 |
ENST00000441339.2
ENST00000308946.3 ENST00000535407.1 |
MYEOV
|
myeloma overexpressed |
| chr2_+_37571717 | 0.29 |
ENST00000338415.3
ENST00000404976.1 |
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr19_-_58609570 | 0.27 |
ENST00000600845.1
ENST00000240727.6 ENST00000600897.1 ENST00000421612.2 ENST00000601063.1 ENST00000601144.1 |
ZSCAN18
|
zinc finger and SCAN domain containing 18 |
| chr19_+_13906250 | 0.27 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr12_-_48152853 | 0.27 |
ENST00000171000.4
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr14_+_24630465 | 0.25 |
ENST00000557894.1
ENST00000559284.1 ENST00000560275.1 |
IRF9
|
interferon regulatory factor 9 |
| chr16_+_50313426 | 0.25 |
ENST00000569265.1
|
ADCY7
|
adenylate cyclase 7 |
| chr16_+_2570340 | 0.24 |
ENST00000568263.1
ENST00000293971.6 ENST00000302956.4 ENST00000413459.3 ENST00000566706.1 ENST00000569879.1 |
AMDHD2
|
amidohydrolase domain containing 2 |
| chr16_-_88717482 | 0.24 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr6_+_32006159 | 0.24 |
ENST00000478281.1
ENST00000471671.1 ENST00000435122.2 |
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr7_+_65540853 | 0.24 |
ENST00000380839.4
ENST00000395332.3 ENST00000362000.5 ENST00000395331.3 |
ASL
|
argininosuccinate lyase |
| chr17_+_45331184 | 0.23 |
ENST00000559488.1
ENST00000571680.1 ENST00000435993.2 |
ITGB3
|
integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) |
| chr1_-_32801825 | 0.23 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr1_+_43148059 | 0.23 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr7_+_150020329 | 0.22 |
ENST00000323078.7
|
LRRC61
|
leucine rich repeat containing 61 |
| chr1_+_180875711 | 0.22 |
ENST00000434447.1
|
RP11-46A10.2
|
RP11-46A10.2 |
| chr7_+_79763271 | 0.21 |
ENST00000442586.1
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr11_-_60719213 | 0.21 |
ENST00000227880.3
|
SLC15A3
|
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr11_+_58938903 | 0.21 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr15_+_73976715 | 0.20 |
ENST00000558689.1
ENST00000560786.2 ENST00000561213.1 ENST00000563584.1 ENST00000561416.1 |
CD276
|
CD276 molecule |
| chr21_-_45196326 | 0.19 |
ENST00000291568.5
|
CSTB
|
cystatin B (stefin B) |
| chr19_-_1174226 | 0.19 |
ENST00000587024.1
ENST00000361757.3 |
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr19_+_17970693 | 0.19 |
ENST00000600147.1
ENST00000599898.1 |
RPL18A
|
ribosomal protein L18a |
| chr2_+_37571845 | 0.18 |
ENST00000537448.1
|
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr7_+_150758642 | 0.18 |
ENST00000488420.1
|
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr10_+_71561630 | 0.18 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr19_+_49617581 | 0.18 |
ENST00000391864.3
|
LIN7B
|
lin-7 homolog B (C. elegans) |
| chr7_+_150758304 | 0.18 |
ENST00000482950.1
ENST00000463414.1 ENST00000310317.5 |
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr9_+_140083099 | 0.18 |
ENST00000322310.5
|
SSNA1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr9_-_140082983 | 0.18 |
ENST00000323927.2
|
ANAPC2
|
anaphase promoting complex subunit 2 |
| chr7_+_150020363 | 0.17 |
ENST00000359623.4
ENST00000493307.1 |
LRRC61
|
leucine rich repeat containing 61 |
| chr11_+_64692143 | 0.17 |
ENST00000164133.2
ENST00000532850.1 |
PPP2R5B
|
protein phosphatase 2, regulatory subunit B', beta |
| chr7_+_65540780 | 0.17 |
ENST00000304874.9
|
ASL
|
argininosuccinate lyase |
| chrX_-_102319092 | 0.17 |
ENST00000372728.3
|
BEX1
|
brain expressed, X-linked 1 |
| chr17_-_4890919 | 0.16 |
ENST00000572543.1
ENST00000381311.5 ENST00000348066.3 ENST00000358183.4 |
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr1_+_3370990 | 0.16 |
ENST00000378378.4
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr17_+_27894180 | 0.16 |
ENST00000583940.1
|
TP53I13
|
tumor protein p53 inducible protein 13 |
| chr1_-_45308616 | 0.16 |
ENST00000447098.2
ENST00000372192.3 |
PTCH2
|
patched 2 |
| chr16_-_74808710 | 0.16 |
ENST00000219368.3
ENST00000544337.1 |
FA2H
|
fatty acid 2-hydroxylase |
| chr16_+_2059872 | 0.16 |
ENST00000567649.1
|
NPW
|
neuropeptide W |
| chr19_-_18717627 | 0.16 |
ENST00000392386.3
|
CRLF1
|
cytokine receptor-like factor 1 |
| chr16_-_2059797 | 0.16 |
ENST00000563630.1
|
ZNF598
|
zinc finger protein 598 |
| chr22_+_43506747 | 0.15 |
ENST00000216115.2
|
BIK
|
BCL2-interacting killer (apoptosis-inducing) |
| chr19_+_17970677 | 0.15 |
ENST00000222247.5
|
RPL18A
|
ribosomal protein L18a |
| chr22_+_29702572 | 0.15 |
ENST00000407647.2
ENST00000416823.1 ENST00000428622.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr19_-_40791302 | 0.15 |
ENST00000392038.2
ENST00000578123.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr1_-_31381528 | 0.15 |
ENST00000339394.6
|
SDC3
|
syndecan 3 |
| chr16_-_29466285 | 0.15 |
ENST00000330978.3
|
BOLA2
|
bolA family member 2 |
| chr11_-_65667997 | 0.15 |
ENST00000312562.2
ENST00000534222.1 |
FOSL1
|
FOS-like antigen 1 |
| chr11_-_65667884 | 0.15 |
ENST00000448083.2
ENST00000531493.1 ENST00000532401.1 |
FOSL1
|
FOS-like antigen 1 |
| chr8_-_145060593 | 0.14 |
ENST00000313059.5
ENST00000524918.1 ENST00000313028.7 ENST00000525773.1 |
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr9_+_116638562 | 0.14 |
ENST00000374126.5
ENST00000288466.7 |
ZNF618
|
zinc finger protein 618 |
| chr20_-_62129163 | 0.14 |
ENST00000298049.7
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr14_+_71108460 | 0.14 |
ENST00000256367.2
|
TTC9
|
tetratricopeptide repeat domain 9 |
| chr16_-_30205627 | 0.14 |
ENST00000305321.4
|
BOLA2B
|
bolA family member 2B |
| chr5_-_176924562 | 0.14 |
ENST00000359895.2
ENST00000355572.2 ENST00000355841.2 ENST00000393551.1 ENST00000505074.1 ENST00000356618.4 ENST00000393546.4 |
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chr9_-_117880477 | 0.14 |
ENST00000534839.1
ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC
|
tenascin C |
| chr18_+_2846972 | 0.14 |
ENST00000254528.3
|
EMILIN2
|
elastin microfibril interfacer 2 |
| chr22_-_39636914 | 0.14 |
ENST00000381551.4
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr19_-_48753028 | 0.14 |
ENST00000522431.1
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr11_+_73675873 | 0.13 |
ENST00000537753.1
ENST00000542350.1 |
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr3_-_49203744 | 0.13 |
ENST00000321895.6
|
CCDC71
|
coiled-coil domain containing 71 |
| chr9_+_132565454 | 0.13 |
ENST00000427860.1
|
TOR1B
|
torsin family 1, member B (torsin B) |
| chr16_+_89989687 | 0.13 |
ENST00000315491.7
ENST00000555576.1 ENST00000554336.1 ENST00000553967.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr1_+_43148625 | 0.13 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chr7_-_92107229 | 0.13 |
ENST00000603053.1
|
ERVW-1
|
endogenous retrovirus group W, member 1 |
| chr16_+_2039946 | 0.13 |
ENST00000248121.2
ENST00000568896.1 |
SYNGR3
|
synaptogyrin 3 |
| chr9_-_139581848 | 0.13 |
ENST00000538402.1
ENST00000371694.3 |
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 |
| chr3_-_87039662 | 0.13 |
ENST00000494229.1
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr20_+_55967129 | 0.13 |
ENST00000371219.2
|
RBM38
|
RNA binding motif protein 38 |
| chr17_-_4871085 | 0.13 |
ENST00000575142.1
ENST00000206020.3 |
SPAG7
|
sperm associated antigen 7 |
| chr3_-_134093395 | 0.12 |
ENST00000249883.5
|
AMOTL2
|
angiomotin like 2 |
| chr17_+_7531281 | 0.12 |
ENST00000575729.1
ENST00000340624.5 |
SHBG
|
sex hormone-binding globulin |
| chr7_+_149571045 | 0.12 |
ENST00000479613.1
ENST00000606024.1 ENST00000464662.1 ENST00000425642.2 |
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2 |
| chr1_+_156024552 | 0.12 |
ENST00000368304.5
ENST00000368302.3 |
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr9_-_98279241 | 0.12 |
ENST00000437951.1
ENST00000375274.2 ENST00000430669.2 ENST00000468211.2 |
PTCH1
|
patched 1 |
| chr15_+_75074410 | 0.12 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase |
| chr19_-_37096139 | 0.12 |
ENST00000585983.1
ENST00000585960.1 ENST00000586115.1 |
ZNF529
|
zinc finger protein 529 |
| chr9_+_131580734 | 0.12 |
ENST00000372642.4
|
ENDOG
|
endonuclease G |
| chr3_-_134093738 | 0.12 |
ENST00000506107.1
|
AMOTL2
|
angiomotin like 2 |
| chr11_-_64014379 | 0.12 |
ENST00000309318.3
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr17_-_6616678 | 0.12 |
ENST00000381074.4
ENST00000293800.6 ENST00000572352.1 ENST00000576323.1 ENST00000573648.1 |
SLC13A5
|
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr12_-_57400227 | 0.12 |
ENST00000300101.2
|
ZBTB39
|
zinc finger and BTB domain containing 39 |
| chr17_-_78194147 | 0.12 |
ENST00000534910.1
ENST00000326317.6 |
SGSH
|
N-sulfoglucosamine sulfohydrolase |
| chr17_+_78194261 | 0.12 |
ENST00000572725.1
|
SLC26A11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr11_-_71159458 | 0.11 |
ENST00000355527.3
|
DHCR7
|
7-dehydrocholesterol reductase |
| chr11_-_66139570 | 0.11 |
ENST00000311161.7
|
SLC29A2
|
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
| chr1_-_151119087 | 0.11 |
ENST00000341697.3
ENST00000368914.3 |
SEMA6C
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr2_+_219187871 | 0.11 |
ENST00000258362.3
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr1_+_2407754 | 0.11 |
ENST00000419816.2
ENST00000378486.3 ENST00000378488.3 ENST00000288766.5 |
PLCH2
|
phospholipase C, eta 2 |
| chr16_+_58497587 | 0.11 |
ENST00000569404.1
ENST00000569539.1 ENST00000564126.1 ENST00000565304.1 ENST00000567667.1 |
NDRG4
|
NDRG family member 4 |
| chr1_+_155293702 | 0.11 |
ENST00000368347.4
|
RUSC1
|
RUN and SH3 domain containing 1 |
| chr16_+_2570431 | 0.11 |
ENST00000563556.1
|
AMDHD2
|
amidohydrolase domain containing 2 |
| chr16_+_4364762 | 0.11 |
ENST00000262366.3
|
GLIS2
|
GLIS family zinc finger 2 |
| chr2_+_85132749 | 0.11 |
ENST00000233143.4
|
TMSB10
|
thymosin beta 10 |
| chr16_-_30204987 | 0.11 |
ENST00000569282.1
ENST00000567436.1 |
BOLA2B
|
bolA family member 2B |
| chr7_-_5570229 | 0.11 |
ENST00000331789.5
|
ACTB
|
actin, beta |
| chr22_+_39493207 | 0.11 |
ENST00000348946.4
ENST00000442487.3 ENST00000421988.2 |
APOBEC3H
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
| chr1_-_159915386 | 0.11 |
ENST00000361509.3
ENST00000368094.1 |
IGSF9
|
immunoglobulin superfamily, member 9 |
| chr9_-_139581875 | 0.11 |
ENST00000371696.2
|
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 |
| chr10_+_71561649 | 0.11 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr17_-_4890649 | 0.11 |
ENST00000361571.5
|
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr1_+_28995258 | 0.11 |
ENST00000361872.4
ENST00000294409.2 |
GMEB1
|
glucocorticoid modulatory element binding protein 1 |
| chr11_-_57004658 | 0.11 |
ENST00000606794.1
|
APLNR
|
apelin receptor |
| chr6_-_10419871 | 0.11 |
ENST00000319516.4
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr19_-_49122384 | 0.10 |
ENST00000550973.1
ENST00000550645.1 ENST00000549273.1 ENST00000552588.1 |
RPL18
|
ribosomal protein L18 |
| chr19_-_2151523 | 0.10 |
ENST00000350812.6
ENST00000355272.6 ENST00000356926.4 ENST00000345016.5 |
AP3D1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr5_-_180018540 | 0.10 |
ENST00000292641.3
|
SCGB3A1
|
secretoglobin, family 3A, member 1 |
| chr11_-_66139617 | 0.10 |
ENST00000544554.1
ENST00000546034.1 |
SLC29A2
|
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
| chr3_+_71803201 | 0.10 |
ENST00000304411.2
|
GPR27
|
G protein-coupled receptor 27 |
| chr11_-_67276100 | 0.10 |
ENST00000301488.3
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr2_+_105760549 | 0.10 |
ENST00000436841.1
ENST00000452037.1 ENST00000438148.1 |
AC104655.3
|
AC104655.3 |
| chr19_+_984313 | 0.10 |
ENST00000251289.5
ENST00000587001.2 ENST00000607440.1 |
WDR18
|
WD repeat domain 18 |
| chr19_+_10362882 | 0.10 |
ENST00000393733.2
ENST00000588502.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr19_+_5823813 | 0.10 |
ENST00000303212.2
|
NRTN
|
neurturin |
| chr19_+_10362577 | 0.10 |
ENST00000592514.1
ENST00000307422.5 ENST00000253099.6 ENST00000590150.1 ENST00000590669.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr14_+_29236269 | 0.10 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr5_+_176731572 | 0.10 |
ENST00000503853.1
|
PRELID1
|
PRELI domain containing 1 |
| chr3_-_129035120 | 0.10 |
ENST00000333762.4
|
H1FX
|
H1 histone family, member X |
| chr5_+_145316120 | 0.10 |
ENST00000359120.4
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr2_-_96811170 | 0.10 |
ENST00000288943.4
|
DUSP2
|
dual specificity phosphatase 2 |
| chr20_+_3776936 | 0.10 |
ENST00000439880.2
|
CDC25B
|
cell division cycle 25B |
| chr17_-_76124812 | 0.10 |
ENST00000592063.1
ENST00000589271.1 ENST00000322933.4 ENST00000589553.1 |
TMC6
|
transmembrane channel-like 6 |
| chr11_-_407103 | 0.10 |
ENST00000526395.1
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr4_-_140477910 | 0.10 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr7_-_150020578 | 0.10 |
ENST00000478393.1
|
ACTR3C
|
ARP3 actin-related protein 3 homolog C (yeast) |
| chr2_-_242626127 | 0.09 |
ENST00000445261.1
|
DTYMK
|
deoxythymidylate kinase (thymidylate kinase) |
| chr17_-_70053866 | 0.09 |
ENST00000540802.1
|
RP11-84E24.2
|
RP11-84E24.2 |
| chr17_-_1083078 | 0.09 |
ENST00000574266.1
ENST00000302538.5 |
ABR
|
active BCR-related |
| chrX_+_105066524 | 0.09 |
ENST00000243300.9
ENST00000428173.2 |
NRK
|
Nik related kinase |
| chr1_+_156024525 | 0.09 |
ENST00000368305.4
|
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr12_+_49932886 | 0.09 |
ENST00000257981.6
|
KCNH3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chrX_+_153686614 | 0.09 |
ENST00000369682.3
|
PLXNA3
|
plexin A3 |
| chr15_+_75074385 | 0.09 |
ENST00000220003.9
|
CSK
|
c-src tyrosine kinase |
| chr17_-_78194124 | 0.09 |
ENST00000570427.1
ENST00000570923.1 |
SGSH
|
N-sulfoglucosamine sulfohydrolase |
| chr19_+_33685490 | 0.09 |
ENST00000253193.7
|
LRP3
|
low density lipoprotein receptor-related protein 3 |
| chr22_-_50683381 | 0.09 |
ENST00000439308.2
|
TUBGCP6
|
tubulin, gamma complex associated protein 6 |
| chr6_+_32006042 | 0.09 |
ENST00000418967.2
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr9_-_34662651 | 0.09 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr19_-_40791211 | 0.09 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr22_-_19466643 | 0.09 |
ENST00000474226.1
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr2_+_219264762 | 0.09 |
ENST00000452977.1
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr17_+_77030267 | 0.09 |
ENST00000581774.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr9_-_123639445 | 0.09 |
ENST00000312189.6
|
PHF19
|
PHD finger protein 19 |
| chr16_-_402639 | 0.09 |
ENST00000262320.3
|
AXIN1
|
axin 1 |
| chr15_+_31619013 | 0.09 |
ENST00000307145.3
|
KLF13
|
Kruppel-like factor 13 |
| chr3_-_9811595 | 0.09 |
ENST00000256460.3
|
CAMK1
|
calcium/calmodulin-dependent protein kinase I |
| chr19_-_1863567 | 0.09 |
ENST00000250916.4
|
KLF16
|
Kruppel-like factor 16 |
| chr17_-_39183452 | 0.09 |
ENST00000361883.5
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr14_+_105886275 | 0.09 |
ENST00000405646.1
|
MTA1
|
metastasis associated 1 |
| chr19_-_50063907 | 0.08 |
ENST00000598296.1
|
NOSIP
|
nitric oxide synthase interacting protein |
| chr14_+_105886150 | 0.08 |
ENST00000331320.7
ENST00000406191.1 |
MTA1
|
metastasis associated 1 |
| chr10_+_6779326 | 0.08 |
ENST00000417112.1
|
RP11-554I8.2
|
RP11-554I8.2 |
| chr16_+_777739 | 0.08 |
ENST00000563792.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr12_-_51419924 | 0.08 |
ENST00000541174.2
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr11_-_71159380 | 0.08 |
ENST00000525346.1
ENST00000531364.1 ENST00000529990.1 ENST00000527316.1 ENST00000407721.2 |
DHCR7
|
7-dehydrocholesterol reductase |
| chr6_-_29595779 | 0.08 |
ENST00000355973.3
ENST00000377012.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr7_+_99775520 | 0.08 |
ENST00000317296.5
ENST00000422690.1 ENST00000439782.1 |
STAG3
|
stromal antigen 3 |
| chr16_+_56691838 | 0.08 |
ENST00000394501.2
|
MT1F
|
metallothionein 1F |
| chr16_+_89753070 | 0.08 |
ENST00000353379.7
ENST00000505473.1 ENST00000564192.1 |
CDK10
|
cyclin-dependent kinase 10 |
| chr1_+_228194693 | 0.08 |
ENST00000284523.1
ENST00000366753.2 |
WNT3A
|
wingless-type MMTV integration site family, member 3A |
| chr19_-_40791160 | 0.08 |
ENST00000358335.5
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr7_-_5569588 | 0.08 |
ENST00000417101.1
|
ACTB
|
actin, beta |
| chrX_+_51927919 | 0.08 |
ENST00000416960.1
|
MAGED4
|
melanoma antigen family D, 4 |
| chr21_+_45725050 | 0.08 |
ENST00000403390.1
|
PFKL
|
phosphofructokinase, liver |
| chr19_-_49314169 | 0.08 |
ENST00000597011.1
ENST00000601681.1 |
BCAT2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr19_-_49314269 | 0.08 |
ENST00000545387.2
ENST00000316273.6 ENST00000402551.1 ENST00000598162.1 ENST00000599246.1 |
BCAT2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr17_-_78194716 | 0.08 |
ENST00000576707.1
|
SGSH
|
N-sulfoglucosamine sulfohydrolase |
| chr20_-_62601218 | 0.08 |
ENST00000369888.1
|
ZNF512B
|
zinc finger protein 512B |
| chr16_+_29818857 | 0.08 |
ENST00000567444.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr2_+_105050794 | 0.08 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr13_-_52378231 | 0.08 |
ENST00000280056.2
ENST00000444610.2 |
DHRS12
|
dehydrogenase/reductase (SDR family) member 12 |
| chr4_-_168155417 | 0.08 |
ENST00000511269.1
ENST00000506697.1 ENST00000512042.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr16_+_58497567 | 0.08 |
ENST00000258187.5
|
NDRG4
|
NDRG family member 4 |
| chr17_+_39421591 | 0.08 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6 |
| chr1_+_157963391 | 0.08 |
ENST00000359209.6
ENST00000416935.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr17_-_79481666 | 0.08 |
ENST00000575659.1
|
ACTG1
|
actin, gamma 1 |
| chr6_-_29600832 | 0.08 |
ENST00000377016.4
ENST00000376977.3 ENST00000377034.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr11_-_96239990 | 0.07 |
ENST00000511243.2
|
JRKL-AS1
|
JRKL antisense RNA 1 |
| chr20_+_3777078 | 0.07 |
ENST00000340833.4
|
CDC25B
|
cell division cycle 25B |
| chr2_+_23608064 | 0.07 |
ENST00000486442.1
|
KLHL29
|
kelch-like family member 29 |
| chr11_-_64545941 | 0.07 |
ENST00000377387.1
|
SF1
|
splicing factor 1 |
| chr16_+_30205225 | 0.07 |
ENST00000345535.4
ENST00000251303.6 |
SLX1A
|
SLX1 structure-specific endonuclease subunit homolog A (S. cerevisiae) |
| chr10_-_131762105 | 0.07 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr2_+_103236004 | 0.07 |
ENST00000233969.2
|
SLC9A2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr2_+_238768187 | 0.07 |
ENST00000254661.4
ENST00000409726.1 |
RAMP1
|
receptor (G protein-coupled) activity modifying protein 1 |
| chr3_+_100053542 | 0.07 |
ENST00000394140.4
|
NIT2
|
nitrilase family, member 2 |
| chr1_+_67773527 | 0.07 |
ENST00000541374.1
ENST00000544434.1 |
IL12RB2
|
interleukin 12 receptor, beta 2 |
| chr1_-_114355083 | 0.07 |
ENST00000261441.5
|
RSBN1
|
round spermatid basic protein 1 |
| chr19_-_31840130 | 0.07 |
ENST00000558569.1
|
TSHZ3
|
teashirt zinc finger homeobox 3 |
| chr11_-_66139199 | 0.07 |
ENST00000357440.2
|
SLC29A2
|
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
| chr15_-_91565743 | 0.07 |
ENST00000535843.1
|
VPS33B
|
vacuolar protein sorting 33 homolog B (yeast) |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC3_ZIC4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 0.4 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.1 | 0.5 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.1 | 0.6 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.1 | 0.1 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.1 | 0.2 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 0.2 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.1 | 0.4 | GO:0071486 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.3 | GO:0033123 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.2 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.2 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.4 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.3 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.2 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.2 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0048007 | synaptic vesicle recycling via endosome(GO:0036466) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.1 | GO:1900241 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.0 | 0.2 | GO:0033489 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.1 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.0 | 0.1 | GO:0048320 | forebrain anterior/posterior pattern specification(GO:0021797) axial mesoderm formation(GO:0048320) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.2 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.0 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.2 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.2 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) optic cup structural organization(GO:0003409) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.0 | 0.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.3 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.1 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.0 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.3 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.0 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.0 | 0.4 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.0 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.1 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.2 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.2 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:2000855 | mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) positive regulation of mineralocorticoid secretion(GO:2000857) regulation of aldosterone secretion(GO:2000858) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.4 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.0 | GO:0051795 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) atrial septum primum morphogenesis(GO:0003289) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) uterine wall breakdown(GO:0042704) positive regulation of catagen(GO:0051795) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.0 | 0.1 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.1 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.0 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.3 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 0.2 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.4 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0019031 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.3 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.0 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.0 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.5 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.1 | 0.3 | GO:0016826 | N-sulfoglucosamine sulfohydrolase activity(GO:0016250) hydrolase activity, acting on acid sulfur-nitrogen bonds(GO:0016826) |
| 0.0 | 0.3 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.4 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.3 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.2 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.4 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.2 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.0 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.0 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.0 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.0 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.6 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |