Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
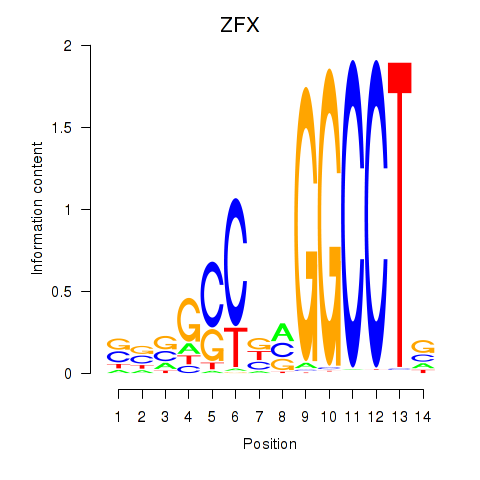
Results for ZFX
Z-value: 1.41
Transcription factors associated with ZFX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZFX
|
ENSG00000005889.11 | zinc finger protein X-linked |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZFX | hg19_v2_chrX_+_24167746_24167811 | 0.56 | 2.4e-01 | Click! |
Activity profile of ZFX motif
Sorted Z-values of ZFX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_144651024 | 1.08 |
ENST00000524906.1
ENST00000532862.1 ENST00000534459.1 |
MROH6
|
maestro heat-like repeat family member 6 |
| chr12_-_122241812 | 0.92 |
ENST00000538335.1
|
AC084018.1
|
AC084018.1 |
| chr19_+_2096868 | 0.63 |
ENST00000395296.1
ENST00000395301.3 |
IZUMO4
|
IZUMO family member 4 |
| chr7_-_1499123 | 0.63 |
ENST00000297508.7
|
MICALL2
|
MICAL-like 2 |
| chr1_-_1850697 | 0.62 |
ENST00000378598.4
ENST00000416272.1 ENST00000310991.3 |
TMEM52
|
transmembrane protein 52 |
| chr7_+_100210133 | 0.61 |
ENST00000393950.2
ENST00000424091.2 |
MOSPD3
|
motile sperm domain containing 3 |
| chr11_-_67397371 | 0.61 |
ENST00000376693.2
ENST00000301490.4 |
NUDT8
|
nudix (nucleoside diphosphate linked moiety X)-type motif 8 |
| chr17_+_21188012 | 0.60 |
ENST00000529517.1
|
MAP2K3
|
mitogen-activated protein kinase kinase 3 |
| chr2_-_160143158 | 0.60 |
ENST00000409124.1
ENST00000358147.4 |
WDSUB1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chr15_+_31658349 | 0.59 |
ENST00000558844.1
|
KLF13
|
Kruppel-like factor 13 |
| chr20_+_47662805 | 0.57 |
ENST00000262982.2
ENST00000542325.1 |
CSE1L
|
CSE1 chromosome segregation 1-like (yeast) |
| chr8_-_145086922 | 0.55 |
ENST00000530478.1
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr4_+_6717842 | 0.55 |
ENST00000320776.3
|
BLOC1S4
|
biogenesis of lysosomal organelles complex-1, subunit 4, cappuccino |
| chr19_-_4065730 | 0.52 |
ENST00000601588.1
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr7_-_1498962 | 0.52 |
ENST00000405088.4
|
MICALL2
|
MICAL-like 2 |
| chr19_+_45844032 | 0.51 |
ENST00000589837.1
|
KLC3
|
kinesin light chain 3 |
| chr7_+_100209979 | 0.51 |
ENST00000493970.1
ENST00000379527.2 |
MOSPD3
|
motile sperm domain containing 3 |
| chr16_-_67260901 | 0.49 |
ENST00000341546.3
ENST00000409509.1 ENST00000433915.1 ENST00000454102.2 |
LRRC29
AC040160.1
|
leucine rich repeat containing 29 Uncharacterized protein; cDNA FLJ57407, weakly similar to Mus musculus leucine rich repeat containing 29 (Lrrc29), mRNA |
| chr12_-_120907374 | 0.49 |
ENST00000550458.1
|
SRSF9
|
serine/arginine-rich splicing factor 9 |
| chr9_+_136325089 | 0.48 |
ENST00000291722.7
ENST00000316948.4 ENST00000540581.1 |
CACFD1
|
calcium channel flower domain containing 1 |
| chr20_-_60942326 | 0.48 |
ENST00000370677.3
ENST00000370692.3 |
LAMA5
|
laminin, alpha 5 |
| chr19_-_8579030 | 0.46 |
ENST00000255616.8
ENST00000393927.4 |
ZNF414
|
zinc finger protein 414 |
| chr2_+_162101247 | 0.45 |
ENST00000439050.1
ENST00000436506.1 |
AC009299.3
|
AC009299.3 |
| chr19_-_1592652 | 0.44 |
ENST00000156825.1
ENST00000434436.3 |
MBD3
|
methyl-CpG binding domain protein 3 |
| chr12_+_132379160 | 0.44 |
ENST00000321867.4
|
ULK1
|
unc-51 like autophagy activating kinase 1 |
| chr10_+_102729249 | 0.44 |
ENST00000519649.1
ENST00000518124.1 |
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G |
| chr16_-_2246436 | 0.44 |
ENST00000343516.6
|
CASKIN1
|
CASK interacting protein 1 |
| chr19_+_13228917 | 0.43 |
ENST00000586171.1
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr7_+_100209725 | 0.43 |
ENST00000223054.4
|
MOSPD3
|
motile sperm domain containing 3 |
| chr12_-_50222187 | 0.43 |
ENST00000335999.6
|
NCKAP5L
|
NCK-associated protein 5-like |
| chr4_-_103748271 | 0.43 |
ENST00000343106.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr19_-_53238260 | 0.42 |
ENST00000453741.2
ENST00000602162.1 ENST00000601643.1 ENST00000596702.1 ENST00000600943.1 ENST00000543227.1 ENST00000540744.1 |
ZNF611
|
zinc finger protein 611 |
| chr2_-_160143059 | 0.42 |
ENST00000392796.3
|
WDSUB1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chr1_-_1284730 | 0.41 |
ENST00000378888.5
|
DVL1
|
dishevelled segment polarity protein 1 |
| chr2_-_20101701 | 0.41 |
ENST00000402414.1
ENST00000333610.3 |
TTC32
|
tetratricopeptide repeat domain 32 |
| chr20_-_60942361 | 0.41 |
ENST00000252999.3
|
LAMA5
|
laminin, alpha 5 |
| chr5_+_56205878 | 0.41 |
ENST00000423328.1
|
SETD9
|
SET domain containing 9 |
| chr19_-_16653325 | 0.40 |
ENST00000546361.2
|
CHERP
|
calcium homeostasis endoplasmic reticulum protein |
| chr4_-_967326 | 0.40 |
ENST00000273814.3
|
DGKQ
|
diacylglycerol kinase, theta 110kDa |
| chr19_+_1383890 | 0.39 |
ENST00000539480.1
ENST00000313408.7 ENST00000414651.2 |
NDUFS7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase) |
| chr19_+_45844018 | 0.38 |
ENST00000585434.1
|
KLC3
|
kinesin light chain 3 |
| chr12_+_122064673 | 0.38 |
ENST00000537188.1
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chr11_+_844067 | 0.38 |
ENST00000397406.1
ENST00000409543.2 ENST00000525201.1 |
TSPAN4
|
tetraspanin 4 |
| chr19_+_50706866 | 0.38 |
ENST00000440075.2
ENST00000376970.2 ENST00000425460.1 ENST00000599920.1 ENST00000601313.1 |
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr11_-_64885111 | 0.38 |
ENST00000528598.1
ENST00000310597.4 |
ZNHIT2
|
zinc finger, HIT-type containing 2 |
| chr7_-_155437075 | 0.37 |
ENST00000401694.1
|
AC009403.2
|
Protein LOC100506302 |
| chr11_+_705193 | 0.37 |
ENST00000527199.1
|
EPS8L2
|
EPS8-like 2 |
| chr19_-_16653226 | 0.37 |
ENST00000198939.6
|
CHERP
|
calcium homeostasis endoplasmic reticulum protein |
| chr17_-_62493131 | 0.37 |
ENST00000539111.2
|
POLG2
|
polymerase (DNA directed), gamma 2, accessory subunit |
| chr6_-_29600559 | 0.37 |
ENST00000476670.1
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr22_+_50639408 | 0.37 |
ENST00000380903.2
|
SELO
|
Selenoprotein O |
| chr11_+_66624527 | 0.37 |
ENST00000393952.3
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr3_+_48507210 | 0.36 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr19_+_1450112 | 0.36 |
ENST00000590469.1
ENST00000233607.2 ENST00000238483.4 ENST00000590877.1 |
APC2
|
adenomatosis polyposis coli 2 |
| chr19_-_19006920 | 0.36 |
ENST00000429504.2
ENST00000427170.2 |
CERS1
|
ceramide synthase 1 |
| chr1_-_85040090 | 0.36 |
ENST00000370630.5
|
CTBS
|
chitobiase, di-N-acetyl- |
| chr19_+_45843994 | 0.36 |
ENST00000391946.2
|
KLC3
|
kinesin light chain 3 |
| chr2_-_160143084 | 0.36 |
ENST00000409990.3
|
WDSUB1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chr15_+_73976715 | 0.35 |
ENST00000558689.1
ENST00000560786.2 ENST00000561213.1 ENST00000563584.1 ENST00000561416.1 |
CD276
|
CD276 molecule |
| chr11_+_66025091 | 0.35 |
ENST00000526758.1
ENST00000440228.2 |
KLC2
|
kinesin light chain 2 |
| chr14_+_64970427 | 0.35 |
ENST00000553583.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr8_-_142318398 | 0.34 |
ENST00000520137.1
|
SLC45A4
|
solute carrier family 45, member 4 |
| chr8_-_101322132 | 0.34 |
ENST00000523481.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr11_+_576494 | 0.33 |
ENST00000533464.1
ENST00000413872.2 ENST00000416188.2 |
PHRF1
|
PHD and ring finger domains 1 |
| chr7_-_127032114 | 0.33 |
ENST00000436992.1
|
ZNF800
|
zinc finger protein 800 |
| chr16_-_88717482 | 0.33 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr22_-_50746027 | 0.33 |
ENST00000425954.1
ENST00000449103.1 |
PLXNB2
|
plexin B2 |
| chr16_+_2034692 | 0.33 |
ENST00000567719.1
|
GFER
|
growth factor, augmenter of liver regeneration |
| chr17_-_73844722 | 0.32 |
ENST00000586257.1
|
WBP2
|
WW domain binding protein 2 |
| chr11_-_507184 | 0.32 |
ENST00000533410.1
ENST00000354420.2 ENST00000397604.3 ENST00000531149.1 ENST00000356187.5 |
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr19_+_13122980 | 0.32 |
ENST00000590027.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr16_+_67563250 | 0.32 |
ENST00000566907.1
|
FAM65A
|
family with sequence similarity 65, member A |
| chr22_-_50970566 | 0.32 |
ENST00000405135.1
ENST00000401779.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr7_-_108210048 | 0.32 |
ENST00000415914.3
ENST00000438865.1 |
THAP5
|
THAP domain containing 5 |
| chr7_+_150076406 | 0.32 |
ENST00000329630.5
|
ZNF775
|
zinc finger protein 775 |
| chr5_+_134074231 | 0.32 |
ENST00000514518.1
|
CAMLG
|
calcium modulating ligand |
| chr11_+_65339820 | 0.31 |
ENST00000316409.2
ENST00000449319.2 ENST00000530349.1 |
FAM89B
|
family with sequence similarity 89, member B |
| chr16_-_1821721 | 0.31 |
ENST00000219302.3
|
NME3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr16_+_67700673 | 0.31 |
ENST00000403458.4
ENST00000602365.1 |
C16orf86
|
chromosome 16 open reading frame 86 |
| chr16_+_68298466 | 0.31 |
ENST00000568088.1
ENST00000564708.1 |
SLC7A6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
| chr16_-_29466285 | 0.31 |
ENST00000330978.3
|
BOLA2
|
bolA family member 2 |
| chr7_+_150811705 | 0.31 |
ENST00000335367.3
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr15_+_44829255 | 0.31 |
ENST00000261868.5
ENST00000424492.3 |
EIF3J
|
eukaryotic translation initiation factor 3, subunit J |
| chr2_-_160143242 | 0.31 |
ENST00000359774.4
|
WDSUB1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chr12_+_122241928 | 0.30 |
ENST00000604567.1
ENST00000542440.1 |
SETD1B
|
SET domain containing 1B |
| chr19_+_51226573 | 0.30 |
ENST00000250340.4
|
CLEC11A
|
C-type lectin domain family 11, member A |
| chr17_-_77813186 | 0.30 |
ENST00000448310.1
ENST00000269397.4 |
CBX4
|
chromobox homolog 4 |
| chr19_+_4343524 | 0.30 |
ENST00000262966.8
ENST00000359935.4 ENST00000599840.1 |
MPND
|
MPN domain containing |
| chr2_-_206950781 | 0.30 |
ENST00000403263.1
|
INO80D
|
INO80 complex subunit D |
| chr16_+_24550857 | 0.30 |
ENST00000568015.1
|
RBBP6
|
retinoblastoma binding protein 6 |
| chr18_-_47013586 | 0.30 |
ENST00000318240.3
ENST00000579820.1 |
C18orf32
|
chromosome 18 open reading frame 32 |
| chr17_-_36413133 | 0.30 |
ENST00000523089.1
ENST00000312412.4 ENST00000520237.1 |
RP11-1407O15.2
|
TBC1 domain family member 3 |
| chr15_-_72767490 | 0.30 |
ENST00000565181.1
|
RP11-1007O24.3
|
RP11-1007O24.3 |
| chr3_+_149530836 | 0.30 |
ENST00000466478.1
ENST00000491086.1 ENST00000467977.1 |
RNF13
|
ring finger protein 13 |
| chr19_-_663277 | 0.29 |
ENST00000292363.5
|
RNF126
|
ring finger protein 126 |
| chr19_+_58987786 | 0.29 |
ENST00000335841.4
|
ZNF446
|
zinc finger protein 446 |
| chr16_-_4466565 | 0.29 |
ENST00000572467.1
ENST00000423908.2 ENST00000572044.1 ENST00000571052.1 |
CORO7-PAM16
CORO7
|
CORO7-PAM16 readthrough coronin 7 |
| chr14_+_105941118 | 0.29 |
ENST00000550577.1
ENST00000538259.2 |
CRIP2
|
cysteine-rich protein 2 |
| chr19_-_1592828 | 0.29 |
ENST00000592012.1
|
MBD3
|
methyl-CpG binding domain protein 3 |
| chr1_+_180875711 | 0.29 |
ENST00000434447.1
|
RP11-46A10.2
|
RP11-46A10.2 |
| chr16_+_765092 | 0.29 |
ENST00000568223.2
|
METRN
|
meteorin, glial cell differentiation regulator |
| chr2_+_206950095 | 0.29 |
ENST00000435627.1
|
AC007383.3
|
AC007383.3 |
| chr12_+_133066137 | 0.29 |
ENST00000434748.2
|
FBRSL1
|
fibrosin-like 1 |
| chr15_+_44829334 | 0.29 |
ENST00000535391.1
|
EIF3J
|
eukaryotic translation initiation factor 3, subunit J |
| chr7_-_73256838 | 0.29 |
ENST00000297873.4
|
WBSCR27
|
Williams Beuren syndrome chromosome region 27 |
| chr6_+_144471643 | 0.29 |
ENST00000367568.4
|
STX11
|
syntaxin 11 |
| chr22_-_50746001 | 0.28 |
ENST00000359337.4
|
PLXNB2
|
plexin B2 |
| chr11_-_73687997 | 0.28 |
ENST00000545212.1
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr11_+_842808 | 0.28 |
ENST00000397397.2
ENST00000397411.2 ENST00000397396.1 |
TSPAN4
|
tetraspanin 4 |
| chr1_+_100731819 | 0.28 |
ENST00000370126.1
|
RTCA
|
RNA 3'-terminal phosphate cyclase |
| chr1_+_1550795 | 0.28 |
ENST00000520777.1
ENST00000357210.4 ENST00000360522.4 ENST00000378710.3 ENST00000355826.5 ENST00000518681.1 ENST00000505820.2 |
MIB2
|
mindbomb E3 ubiquitin protein ligase 2 |
| chr9_+_35605274 | 0.28 |
ENST00000336395.5
|
TESK1
|
testis-specific kinase 1 |
| chr17_-_74497432 | 0.28 |
ENST00000590288.1
ENST00000313080.4 ENST00000592123.1 ENST00000591255.1 ENST00000585989.1 ENST00000591697.1 ENST00000389760.4 |
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila) |
| chr2_+_67624430 | 0.28 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr9_+_140119618 | 0.28 |
ENST00000359069.2
|
C9orf169
|
chromosome 9 open reading frame 169 |
| chr12_-_57082060 | 0.27 |
ENST00000448157.2
ENST00000414274.3 ENST00000262033.6 ENST00000456859.2 |
PTGES3
|
prostaglandin E synthase 3 (cytosolic) |
| chr19_+_49128209 | 0.27 |
ENST00000599748.1
ENST00000443164.1 ENST00000599029.1 |
SPHK2
|
sphingosine kinase 2 |
| chr11_-_506739 | 0.27 |
ENST00000529306.1
ENST00000438658.2 ENST00000527485.1 ENST00000397615.2 ENST00000397614.1 |
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr5_+_162864575 | 0.27 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr9_+_140317802 | 0.27 |
ENST00000341349.2
ENST00000392815.2 |
NOXA1
|
NADPH oxidase activator 1 |
| chr8_-_144952631 | 0.27 |
ENST00000525985.1
|
EPPK1
|
epiplakin 1 |
| chr19_-_19739321 | 0.27 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr20_+_55967129 | 0.27 |
ENST00000371219.2
|
RBM38
|
RNA binding motif protein 38 |
| chr20_+_49348109 | 0.26 |
ENST00000396039.1
|
PARD6B
|
par-6 family cell polarity regulator beta |
| chr19_+_40697514 | 0.26 |
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr11_+_66824276 | 0.26 |
ENST00000308831.2
|
RHOD
|
ras homolog family member D |
| chrX_+_24167746 | 0.26 |
ENST00000428571.1
ENST00000539115.1 |
ZFX
|
zinc finger protein, X-linked |
| chr12_-_121973974 | 0.26 |
ENST00000538379.1
ENST00000541318.1 ENST00000541511.1 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr16_-_88772670 | 0.26 |
ENST00000562544.1
|
RNF166
|
ring finger protein 166 |
| chr16_-_58033762 | 0.26 |
ENST00000299237.2
|
ZNF319
|
zinc finger protein 319 |
| chr11_+_576457 | 0.26 |
ENST00000264555.5
|
PHRF1
|
PHD and ring finger domains 1 |
| chr3_+_53195517 | 0.26 |
ENST00000487897.1
|
PRKCD
|
protein kinase C, delta |
| chrX_+_153686614 | 0.25 |
ENST00000369682.3
|
PLXNA3
|
plexin A3 |
| chr11_+_842928 | 0.25 |
ENST00000397408.1
|
TSPAN4
|
tetraspanin 4 |
| chr9_+_103235365 | 0.25 |
ENST00000374879.4
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr17_-_76128488 | 0.25 |
ENST00000322914.3
|
TMC6
|
transmembrane channel-like 6 |
| chr12_-_113909877 | 0.25 |
ENST00000261731.3
|
LHX5
|
LIM homeobox 5 |
| chr11_-_67236691 | 0.25 |
ENST00000544903.1
ENST00000308022.2 ENST00000393877.3 ENST00000452789.2 |
TMEM134
|
transmembrane protein 134 |
| chr19_+_41119794 | 0.25 |
ENST00000593463.1
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr17_-_80056099 | 0.25 |
ENST00000306749.2
|
FASN
|
fatty acid synthase |
| chr20_+_43992094 | 0.25 |
ENST00000453003.1
|
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr9_-_6007787 | 0.25 |
ENST00000399933.3
ENST00000381461.2 ENST00000513355.2 |
KIAA2026
|
KIAA2026 |
| chr16_+_3019309 | 0.25 |
ENST00000576565.1
|
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr1_-_1167411 | 0.25 |
ENST00000263741.7
|
SDF4
|
stromal cell derived factor 4 |
| chr16_+_2034183 | 0.24 |
ENST00000569451.1
ENST00000248114.6 ENST00000561710.1 |
GFER
|
growth factor, augmenter of liver regeneration |
| chr9_+_130159504 | 0.24 |
ENST00000373352.1
ENST00000373360.3 |
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr6_-_31612808 | 0.24 |
ENST00000438149.1
|
BAG6
|
BCL2-associated athanogene 6 |
| chr22_+_38321840 | 0.24 |
ENST00000454685.1
|
MICALL1
|
MICAL-like 1 |
| chr1_-_11714700 | 0.24 |
ENST00000354287.4
|
FBXO2
|
F-box protein 2 |
| chrX_+_153030941 | 0.24 |
ENST00000538282.1
|
PLXNB3
|
plexin B3 |
| chr17_-_27916555 | 0.24 |
ENST00000394869.3
|
GIT1
|
G protein-coupled receptor kinase interacting ArfGAP 1 |
| chr5_-_134369973 | 0.24 |
ENST00000265340.7
|
PITX1
|
paired-like homeodomain 1 |
| chr1_-_247171347 | 0.23 |
ENST00000339986.7
ENST00000487338.2 |
ZNF695
|
zinc finger protein 695 |
| chr16_+_28874345 | 0.23 |
ENST00000566209.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr1_-_6453399 | 0.23 |
ENST00000608083.1
|
ACOT7
|
acyl-CoA thioesterase 7 |
| chr19_+_39390320 | 0.23 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr15_-_85197501 | 0.23 |
ENST00000434634.2
|
WDR73
|
WD repeat domain 73 |
| chr22_+_24236191 | 0.23 |
ENST00000215754.7
|
MIF
|
macrophage migration inhibitory factor (glycosylation-inhibiting factor) |
| chr8_+_95731904 | 0.23 |
ENST00000522422.1
|
DPY19L4
|
dpy-19-like 4 (C. elegans) |
| chr1_+_42922173 | 0.23 |
ENST00000455780.1
ENST00000372560.3 ENST00000372561.3 ENST00000372556.3 |
PPCS
|
phosphopantothenoylcysteine synthetase |
| chr11_-_64684672 | 0.23 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr17_-_27916589 | 0.23 |
ENST00000579937.1
ENST00000335356.7 |
GIT1
|
G protein-coupled receptor kinase interacting ArfGAP 1 |
| chr18_-_54305658 | 0.23 |
ENST00000586262.1
ENST00000217515.6 |
TXNL1
|
thioredoxin-like 1 |
| chr12_+_121163538 | 0.23 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr16_+_730379 | 0.23 |
ENST00000567173.1
|
STUB1
|
STIP1 homology and U-box containing protein 1, E3 ubiquitin protein ligase |
| chr15_+_23810903 | 0.23 |
ENST00000564592.1
|
MKRN3
|
makorin ring finger protein 3 |
| chr19_+_19144384 | 0.23 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr2_-_169746878 | 0.23 |
ENST00000282074.2
|
SPC25
|
SPC25, NDC80 kinetochore complex component |
| chr12_+_6898674 | 0.23 |
ENST00000541982.1
ENST00000539492.1 |
CD4
|
CD4 molecule |
| chr4_-_2264015 | 0.23 |
ENST00000337190.2
|
MXD4
|
MAX dimerization protein 4 |
| chr5_+_174151536 | 0.22 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr7_-_138720763 | 0.22 |
ENST00000275766.1
|
ZC3HAV1L
|
zinc finger CCCH-type, antiviral 1-like |
| chr1_+_895930 | 0.22 |
ENST00000338591.3
|
KLHL17
|
kelch-like family member 17 |
| chr9_+_137218362 | 0.22 |
ENST00000481739.1
|
RXRA
|
retinoid X receptor, alpha |
| chr8_-_124253576 | 0.22 |
ENST00000276704.4
|
C8orf76
|
chromosome 8 open reading frame 76 |
| chr16_+_730063 | 0.22 |
ENST00000565677.1
ENST00000219548.4 |
STUB1
|
STIP1 homology and U-box containing protein 1, E3 ubiquitin protein ligase |
| chr3_-_129035120 | 0.22 |
ENST00000333762.4
|
H1FX
|
H1 histone family, member X |
| chr20_+_44462749 | 0.22 |
ENST00000372541.1
|
SNX21
|
sorting nexin family member 21 |
| chr16_+_57139933 | 0.22 |
ENST00000566259.1
|
CPNE2
|
copine II |
| chr7_+_98972327 | 0.22 |
ENST00000455009.1
|
ARPC1B
|
actin related protein 2/3 complex, subunit 1B, 41kDa |
| chr11_+_844406 | 0.22 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr20_-_1165319 | 0.22 |
ENST00000429036.1
|
TMEM74B
|
transmembrane protein 74B |
| chr19_+_13229126 | 0.22 |
ENST00000292431.4
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr5_-_93447333 | 0.21 |
ENST00000395965.3
ENST00000505869.1 ENST00000509163.1 |
FAM172A
|
family with sequence similarity 172, member A |
| chr11_+_65686952 | 0.21 |
ENST00000527119.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr19_+_35491330 | 0.21 |
ENST00000411896.2
ENST00000424536.2 |
GRAMD1A
|
GRAM domain containing 1A |
| chr19_+_12902289 | 0.21 |
ENST00000302754.4
|
JUNB
|
jun B proto-oncogene |
| chr16_-_88772761 | 0.21 |
ENST00000567844.1
ENST00000312838.4 |
RNF166
|
ring finger protein 166 |
| chr19_+_10222189 | 0.21 |
ENST00000321826.4
|
P2RY11
|
purinergic receptor P2Y, G-protein coupled, 11 |
| chr5_-_108063949 | 0.21 |
ENST00000606054.1
|
LINC01023
|
long intergenic non-protein coding RNA 1023 |
| chr16_+_88772866 | 0.21 |
ENST00000453996.2
ENST00000312060.5 ENST00000378384.3 ENST00000567949.1 ENST00000564921.1 |
CTU2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe) |
| chr11_+_66025167 | 0.21 |
ENST00000394067.2
ENST00000316924.5 ENST00000421552.1 ENST00000394078.1 |
KLC2
|
kinesin light chain 2 |
| chr9_-_139922631 | 0.21 |
ENST00000341511.6
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr16_-_1821496 | 0.21 |
ENST00000564628.1
ENST00000563498.1 |
NME3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr22_-_50946113 | 0.21 |
ENST00000216080.5
ENST00000474879.2 ENST00000380796.3 |
LMF2
|
lipase maturation factor 2 |
| chr12_-_111126910 | 0.21 |
ENST00000242607.8
|
HVCN1
|
hydrogen voltage-gated channel 1 |
| chr5_-_137090028 | 0.21 |
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr10_+_22610124 | 0.21 |
ENST00000376663.3
|
BMI1
|
BMI1 polycomb ring finger oncogene |
| chr1_+_44401479 | 0.21 |
ENST00000438616.3
|
ARTN
|
artemin |
| chr19_+_4969116 | 0.21 |
ENST00000588337.1
ENST00000159111.4 ENST00000381759.4 |
KDM4B
|
lysine (K)-specific demethylase 4B |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZFX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.2 | 0.5 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.1 | 0.1 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 0.4 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 0.1 | 0.4 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 0.2 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.1 | 0.6 | GO:0010847 | regulation of chromatin assembly(GO:0010847) protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.4 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.4 | GO:0036114 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 | 0.4 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.1 | 0.5 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 0.1 | GO:1901655 | cellular response to ketone(GO:1901655) |
| 0.1 | 0.8 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.3 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.3 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.5 | GO:0090035 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.1 | 0.4 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.8 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.1 | 0.2 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 0.3 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.1 | 0.3 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.1 | 0.3 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.1 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.3 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.3 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.1 | 0.2 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.1 | 0.2 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.1 | 0.2 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.1 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.2 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 0.2 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.1 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.0 | 0.1 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.2 | GO:1905123 | regulation of glucosylceramidase activity(GO:1905123) |
| 0.0 | 0.7 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.2 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.2 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.3 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.4 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.4 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.2 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.6 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.2 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.3 | GO:1903401 | lysine transport(GO:0015819) L-lysine transport(GO:1902022) L-lysine transmembrane transport(GO:1903401) |
| 0.0 | 0.1 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.4 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.0 | 0.0 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.0 | 0.3 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.4 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.1 | GO:1902512 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.0 | 0.1 | GO:0071440 | regulation of histone H3-K14 acetylation(GO:0071440) |
| 0.0 | 0.1 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.1 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.3 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.1 | GO:1903939 | regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.4 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.0 | 0.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.7 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.8 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.2 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.2 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.0 | 0.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:1902958 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.0 | 0.2 | GO:0071873 | response to norepinephrine(GO:0071873) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.2 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.4 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 1.0 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.0 | 0.4 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.0 | GO:0097485 | neuron projection guidance(GO:0097485) |
| 0.0 | 0.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.2 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:0033029 | neutrophil apoptotic process(GO:0001781) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.0 | 0.2 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.0 | 1.0 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.6 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.4 | GO:0032606 | regulation of type I interferon production(GO:0032479) type I interferon production(GO:0032606) |
| 0.0 | 0.2 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.1 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.2 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.6 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.1 | GO:0044417 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.0 | 0.2 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.2 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.4 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.0 | GO:0071373 | cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.2 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.1 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.2 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) |
| 0.0 | 0.1 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.4 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.1 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.0 | 0.4 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.2 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.1 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.1 | GO:0015827 | tryptophan transport(GO:0015827) |
| 0.0 | 0.2 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.1 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.1 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.1 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.0 | GO:0003134 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 0.0 | 0.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.2 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.3 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.1 | GO:2000543 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.3 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 0.3 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.0 | GO:1901383 | negative regulation of chorionic trophoblast cell proliferation(GO:1901383) |
| 0.0 | 0.4 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.1 | GO:0010160 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.1 | GO:0033591 | response to L-ascorbic acid(GO:0033591) activation of protein kinase C activity(GO:1990051) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.1 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.0 | 0.3 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.1 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.1 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0032796 | uropod organization(GO:0032796) |
| 0.0 | 0.3 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 1.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.0 | 0.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.0 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.0 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.2 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 0.2 | GO:0090045 | positive regulation of deacetylase activity(GO:0090045) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.4 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.0 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.0 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.2 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.1 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.0 | 0.2 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.1 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.3 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.0 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.0 | 0.1 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.0 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.5 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.0 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.1 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0014887 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.1 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.4 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.1 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.2 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.0 | GO:0072134 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) nephrogenic mesenchyme morphogenesis(GO:0072134) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.1 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.0 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.0 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.3 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.0 | GO:1901291 | negative regulation of double-strand break repair via single-strand annealing(GO:1901291) |
| 0.0 | 0.4 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.2 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.0 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.0 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.4 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0021997 | neural plate axis specification(GO:0021997) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.1 | 0.5 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.5 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.6 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.1 | 0.8 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.4 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.1 | 1.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.2 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.1 | 0.3 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 0.3 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.0 | 0.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.6 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 1.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0070993 | translation preinitiation complex(GO:0070993) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.4 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.2 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.0 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.1 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.3 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.5 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.3 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 0.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.5 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.3 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.2 | GO:0031177 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) S-acetyltransferase activity(GO:0016418) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.3 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.4 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.1 | 0.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 1.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.2 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.1 | 0.8 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.1 | 0.2 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 0.1 | 0.2 | GO:0070362 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.1 | 0.2 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.3 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.4 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.3 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 0.2 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.1 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.2 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.2 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 1.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.2 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.2 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.3 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.2 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.1 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.2 | GO:0052812 | phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.1 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.0 | 0.4 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.7 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.3 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.5 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.2 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.6 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.3 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.0 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 1.0 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.0 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.0 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.0 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.0 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 2.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.0 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.2 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.0 | 0.0 | GO:0034039 | 8-oxo-7,8-dihydroguanine DNA N-glycosylase activity(GO:0034039) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.1 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.8 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.8 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |