Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
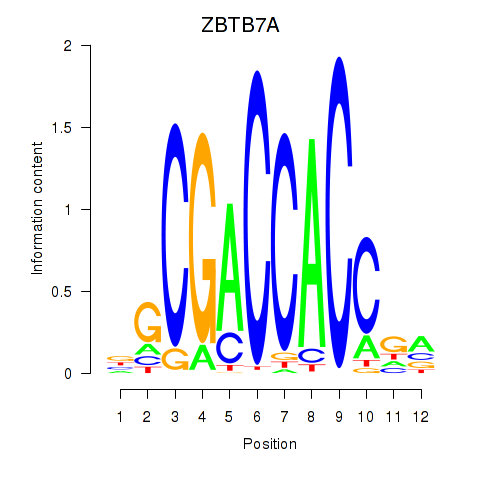
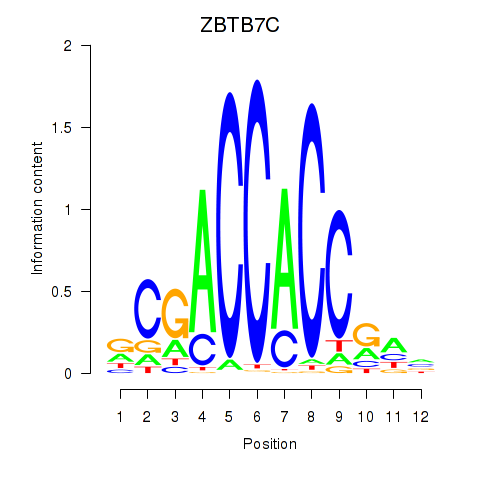
Results for ZBTB7A_ZBTB7C
Z-value: 0.46
Transcription factors associated with ZBTB7A_ZBTB7C
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB7A
|
ENSG00000178951.4 | zinc finger and BTB domain containing 7A |
|
ZBTB7C
|
ENSG00000184828.5 | zinc finger and BTB domain containing 7C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB7A | hg19_v2_chr19_-_4066890_4066943 | 0.62 | 1.9e-01 | Click! |
| ZBTB7C | hg19_v2_chr18_-_45935663_45935793 | -0.11 | 8.3e-01 | Click! |
Activity profile of ZBTB7A_ZBTB7C motif
Sorted Z-values of ZBTB7A_ZBTB7C motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_129872672 | 0.30 |
ENST00000531431.1
ENST00000527581.1 |
PRDM10
|
PR domain containing 10 |
| chr16_+_53164956 | 0.22 |
ENST00000563410.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr21_+_38338737 | 0.20 |
ENST00000430068.1
|
AP000704.5
|
AP000704.5 |
| chr19_-_48673465 | 0.18 |
ENST00000598938.1
|
LIG1
|
ligase I, DNA, ATP-dependent |
| chr5_+_443280 | 0.16 |
ENST00000508022.1
|
EXOC3
|
exocyst complex component 3 |
| chr2_-_235405168 | 0.16 |
ENST00000339728.3
|
ARL4C
|
ADP-ribosylation factor-like 4C |
| chr2_-_37551846 | 0.16 |
ENST00000443187.1
|
PRKD3
|
protein kinase D3 |
| chr4_-_56412713 | 0.15 |
ENST00000435527.2
|
CLOCK
|
clock circadian regulator |
| chr9_+_74526384 | 0.14 |
ENST00000334731.2
ENST00000377031.3 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr5_+_2752216 | 0.13 |
ENST00000457752.2
|
C5orf38
|
chromosome 5 open reading frame 38 |
| chr7_-_127672146 | 0.13 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr19_+_35634146 | 0.13 |
ENST00000586063.1
ENST00000270310.2 ENST00000588265.1 |
FXYD7
|
FXYD domain containing ion transport regulator 7 |
| chr17_+_79989937 | 0.11 |
ENST00000580965.1
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr7_-_127671674 | 0.11 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr14_-_54955721 | 0.10 |
ENST00000554908.1
|
GMFB
|
glia maturation factor, beta |
| chr3_+_108308559 | 0.10 |
ENST00000486815.1
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr15_-_64126084 | 0.10 |
ENST00000560316.1
ENST00000443617.2 ENST00000560462.1 ENST00000558532.1 ENST00000561400.1 |
HERC1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr17_+_79990058 | 0.10 |
ENST00000584341.1
|
RAC3
|
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chr1_+_205538105 | 0.10 |
ENST00000367147.4
ENST00000539267.1 |
MFSD4
|
major facilitator superfamily domain containing 4 |
| chr7_+_26191809 | 0.10 |
ENST00000056233.3
|
NFE2L3
|
nuclear factor, erythroid 2-like 3 |
| chr9_-_123476612 | 0.09 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr17_-_39728303 | 0.09 |
ENST00000588431.1
ENST00000246662.4 |
KRT9
|
keratin 9 |
| chr2_-_97405775 | 0.09 |
ENST00000264963.4
ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L
|
lectin, mannose-binding 2-like |
| chr11_+_48002279 | 0.09 |
ENST00000534219.1
ENST00000527952.1 |
PTPRJ
|
protein tyrosine phosphatase, receptor type, J |
| chr15_-_62352659 | 0.08 |
ENST00000249837.3
|
VPS13C
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr1_-_167906277 | 0.08 |
ENST00000271373.4
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr19_-_5567842 | 0.08 |
ENST00000587632.1
|
TINCR
|
tissue differentiation-inducing non-protein coding RNA |
| chr12_+_57854274 | 0.08 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr7_+_77167343 | 0.08 |
ENST00000433369.2
ENST00000415482.2 |
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr3_+_179040767 | 0.08 |
ENST00000496856.1
ENST00000491818.1 |
ZNF639
|
zinc finger protein 639 |
| chr16_+_53164833 | 0.08 |
ENST00000564845.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr6_-_166755995 | 0.07 |
ENST00000361731.3
|
SFT2D1
|
SFT2 domain containing 1 |
| chr18_-_9614863 | 0.07 |
ENST00000584074.1
|
PPP4R1
|
protein phosphatase 4, regulatory subunit 1 |
| chr11_+_66115304 | 0.07 |
ENST00000531602.1
|
RP11-867G23.8
|
Uncharacterized protein |
| chr5_+_102455968 | 0.07 |
ENST00000358359.3
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr10_+_60144782 | 0.07 |
ENST00000487519.1
|
TFAM
|
transcription factor A, mitochondrial |
| chr14_+_64970662 | 0.07 |
ENST00000556965.1
ENST00000554015.1 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr3_-_71803917 | 0.07 |
ENST00000421769.2
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr11_-_66115032 | 0.06 |
ENST00000311181.4
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 |
| chr6_+_64282447 | 0.06 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr15_-_62352570 | 0.06 |
ENST00000261517.5
ENST00000395896.4 ENST00000395898.3 |
VPS13C
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr20_+_35201993 | 0.06 |
ENST00000373872.4
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr1_+_15480197 | 0.06 |
ENST00000400796.3
ENST00000434578.2 ENST00000376008.2 |
TMEM51
|
transmembrane protein 51 |
| chr1_-_84972248 | 0.06 |
ENST00000370645.4
ENST00000370641.3 |
GNG5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr4_+_74718906 | 0.06 |
ENST00000226524.3
|
PF4V1
|
platelet factor 4 variant 1 |
| chr20_+_44034676 | 0.06 |
ENST00000372723.3
ENST00000372722.3 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr9_+_135037334 | 0.06 |
ENST00000393229.3
ENST00000360670.3 ENST00000393228.4 ENST00000372179.3 |
NTNG2
|
netrin G2 |
| chr4_-_76598296 | 0.06 |
ENST00000395719.3
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chrX_-_128977364 | 0.06 |
ENST00000371064.3
|
ZDHHC9
|
zinc finger, DHHC-type containing 9 |
| chr2_+_46769798 | 0.06 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr8_-_101963482 | 0.06 |
ENST00000419477.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr17_-_5372229 | 0.06 |
ENST00000433302.3
|
DHX33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr7_+_77167376 | 0.06 |
ENST00000435495.2
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr14_-_54955376 | 0.06 |
ENST00000553333.1
|
GMFB
|
glia maturation factor, beta |
| chrX_+_133507327 | 0.06 |
ENST00000332070.3
ENST00000394292.1 ENST00000370799.1 ENST00000416404.2 |
PHF6
|
PHD finger protein 6 |
| chr11_-_129872712 | 0.05 |
ENST00000358825.5
ENST00000360871.3 ENST00000528746.1 |
PRDM10
|
PR domain containing 10 |
| chr8_+_26240414 | 0.05 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr14_+_64970427 | 0.05 |
ENST00000553583.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr5_-_132113690 | 0.05 |
ENST00000414594.1
|
SEPT8
|
septin 8 |
| chr2_+_27346666 | 0.05 |
ENST00000316470.4
ENST00000416071.1 |
ABHD1
|
abhydrolase domain containing 1 |
| chr19_+_10197463 | 0.05 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr12_-_51611477 | 0.05 |
ENST00000389243.4
|
POU6F1
|
POU class 6 homeobox 1 |
| chr12_+_50355647 | 0.05 |
ENST00000293599.6
|
AQP5
|
aquaporin 5 |
| chr2_+_37311588 | 0.05 |
ENST00000409774.1
ENST00000608836.1 |
GPATCH11
|
G patch domain containing 11 |
| chr3_-_179322436 | 0.05 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr17_-_5372068 | 0.05 |
ENST00000572490.1
|
DHX33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr5_+_49963239 | 0.05 |
ENST00000505554.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr1_-_167905225 | 0.05 |
ENST00000367846.4
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr14_+_59951161 | 0.05 |
ENST00000261247.9
ENST00000425728.2 ENST00000556985.1 ENST00000554271.1 ENST00000554795.1 |
JKAMP
|
JNK1/MAPK8-associated membrane protein |
| chr17_+_29158962 | 0.05 |
ENST00000321990.4
|
ATAD5
|
ATPase family, AAA domain containing 5 |
| chr13_+_20532900 | 0.05 |
ENST00000382871.2
|
ZMYM2
|
zinc finger, MYM-type 2 |
| chr1_-_156721502 | 0.05 |
ENST00000357325.5
|
HDGF
|
hepatoma-derived growth factor |
| chr4_-_47916543 | 0.05 |
ENST00000507489.1
|
NFXL1
|
nuclear transcription factor, X-box binding-like 1 |
| chr9_-_77643307 | 0.05 |
ENST00000376834.3
ENST00000376830.3 |
C9orf41
|
chromosome 9 open reading frame 41 |
| chr4_-_76598326 | 0.05 |
ENST00000503660.1
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chrX_+_133507283 | 0.05 |
ENST00000370803.3
|
PHF6
|
PHD finger protein 6 |
| chrX_-_119603138 | 0.05 |
ENST00000200639.4
ENST00000371335.4 ENST00000538785.1 ENST00000434600.2 |
LAMP2
|
lysosomal-associated membrane protein 2 |
| chr4_-_83719884 | 0.05 |
ENST00000282709.4
ENST00000273908.4 |
SCD5
|
stearoyl-CoA desaturase 5 |
| chr3_-_72496035 | 0.05 |
ENST00000477973.2
|
RYBP
|
RING1 and YY1 binding protein |
| chr1_+_162467595 | 0.05 |
ENST00000538489.1
ENST00000489294.1 |
UHMK1
|
U2AF homology motif (UHM) kinase 1 |
| chr5_+_138210919 | 0.05 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr20_+_44034804 | 0.04 |
ENST00000357275.2
ENST00000372720.3 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr15_+_36871983 | 0.04 |
ENST00000437989.2
ENST00000569302.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr17_+_46185111 | 0.04 |
ENST00000582104.1
ENST00000584335.1 |
SNX11
|
sorting nexin 11 |
| chr1_-_111506562 | 0.04 |
ENST00000485275.2
ENST00000369763.4 |
LRIF1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr17_-_56065540 | 0.04 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr10_+_60145155 | 0.04 |
ENST00000373895.3
|
TFAM
|
transcription factor A, mitochondrial |
| chr1_-_156721389 | 0.04 |
ENST00000537739.1
|
HDGF
|
hepatoma-derived growth factor |
| chr8_-_101963677 | 0.04 |
ENST00000395956.3
ENST00000395953.2 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr5_-_2751762 | 0.04 |
ENST00000302057.5
ENST00000382611.6 |
IRX2
|
iroquois homeobox 2 |
| chr3_-_179322416 | 0.04 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr2_-_152118352 | 0.04 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr8_+_22298767 | 0.04 |
ENST00000522000.1
|
PPP3CC
|
protein phosphatase 3, catalytic subunit, gamma isozyme |
| chr19_-_58874112 | 0.04 |
ENST00000311044.3
ENST00000595763.1 ENST00000425453.3 |
ZNF497
|
zinc finger protein 497 |
| chr17_-_30185971 | 0.04 |
ENST00000378634.2
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chr20_-_23402028 | 0.04 |
ENST00000398425.3
ENST00000432543.2 ENST00000377026.4 |
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta |
| chr4_-_76598052 | 0.04 |
ENST00000509100.1
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr2_+_149402553 | 0.04 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr6_-_109703634 | 0.03 |
ENST00000324953.5
ENST00000310786.4 ENST00000275080.7 ENST00000413644.2 |
CD164
|
CD164 molecule, sialomucin |
| chr3_+_179322573 | 0.03 |
ENST00000493866.1
ENST00000472629.1 ENST00000482604.1 |
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr1_+_167905894 | 0.03 |
ENST00000367843.3
ENST00000432587.2 ENST00000312263.6 |
DCAF6
|
DDB1 and CUL4 associated factor 6 |
| chr19_-_663147 | 0.03 |
ENST00000606702.1
|
RNF126
|
ring finger protein 126 |
| chr14_-_61116168 | 0.03 |
ENST00000247182.6
|
SIX1
|
SIX homeobox 1 |
| chr12_+_107349497 | 0.03 |
ENST00000548125.1
ENST00000280756.4 |
C12orf23
|
chromosome 12 open reading frame 23 |
| chr17_-_27044903 | 0.03 |
ENST00000395245.3
|
RAB34
|
RAB34, member RAS oncogene family |
| chr17_+_61600682 | 0.03 |
ENST00000581784.1
ENST00000456941.2 ENST00000580652.1 ENST00000314672.5 ENST00000583023.1 |
KCNH6
|
potassium voltage-gated channel, subfamily H (eag-related), member 6 |
| chr8_+_27629459 | 0.03 |
ENST00000523566.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr7_-_106301405 | 0.03 |
ENST00000523505.1
|
CCDC71L
|
coiled-coil domain containing 71-like |
| chr17_-_39769005 | 0.03 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr9_-_127905736 | 0.03 |
ENST00000336505.6
ENST00000373549.4 |
SCAI
|
suppressor of cancer cell invasion |
| chr20_-_61493115 | 0.03 |
ENST00000335351.3
ENST00000217162.5 |
TCFL5
|
transcription factor-like 5 (basic helix-loop-helix) |
| chr5_+_175792459 | 0.03 |
ENST00000310389.5
|
ARL10
|
ADP-ribosylation factor-like 10 |
| chr11_-_17035943 | 0.03 |
ENST00000355661.3
ENST00000532079.1 ENST00000448080.2 ENST00000531066.1 |
PLEKHA7
|
pleckstrin homology domain containing, family A member 7 |
| chr2_-_153032484 | 0.03 |
ENST00000263904.4
|
STAM2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
| chr5_-_132113559 | 0.03 |
ENST00000448933.1
|
SEPT8
|
septin 8 |
| chr17_-_40428359 | 0.03 |
ENST00000293328.3
|
STAT5B
|
signal transducer and activator of transcription 5B |
| chr10_+_14880364 | 0.03 |
ENST00000441647.1
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr11_+_48002076 | 0.03 |
ENST00000418331.2
ENST00000440289.2 |
PTPRJ
|
protein tyrosine phosphatase, receptor type, J |
| chr8_-_74884482 | 0.03 |
ENST00000520242.1
ENST00000519082.1 |
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr2_+_47630255 | 0.03 |
ENST00000406134.1
|
MSH2
|
mutS homolog 2 |
| chr17_-_39661849 | 0.03 |
ENST00000246635.3
ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13
|
keratin 13 |
| chr6_-_109703600 | 0.03 |
ENST00000512821.1
|
CD164
|
CD164 molecule, sialomucin |
| chr14_-_22005197 | 0.03 |
ENST00000541965.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr17_+_40834580 | 0.02 |
ENST00000264638.4
|
CNTNAP1
|
contactin associated protein 1 |
| chr1_-_156722015 | 0.02 |
ENST00000368209.5
|
HDGF
|
hepatoma-derived growth factor |
| chr3_+_23986748 | 0.02 |
ENST00000312521.4
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr3_-_100120223 | 0.02 |
ENST00000284320.5
|
TOMM70A
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr6_-_17706618 | 0.02 |
ENST00000262077.2
ENST00000537253.1 |
NUP153
|
nucleoporin 153kDa |
| chr5_+_149109825 | 0.02 |
ENST00000360453.4
ENST00000394320.3 ENST00000309241.5 |
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr2_+_172864490 | 0.02 |
ENST00000315796.4
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr18_+_74207477 | 0.02 |
ENST00000532511.1
|
RP11-17M16.1
|
uncharacterized protein LOC400658 |
| chr17_-_27044810 | 0.02 |
ENST00000395242.2
|
RAB34
|
RAB34, member RAS oncogene family |
| chr13_+_20532848 | 0.02 |
ENST00000382874.2
|
ZMYM2
|
zinc finger, MYM-type 2 |
| chr10_+_70661014 | 0.02 |
ENST00000373585.3
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr2_+_47630108 | 0.02 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
| chr12_-_106641728 | 0.02 |
ENST00000378026.4
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr11_-_14380664 | 0.02 |
ENST00000545643.1
ENST00000256196.4 |
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr9_+_74764340 | 0.02 |
ENST00000376986.1
ENST00000358399.3 |
GDA
|
guanine deaminase |
| chr3_-_185542761 | 0.02 |
ENST00000457616.2
ENST00000346192.3 |
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr14_+_59655369 | 0.02 |
ENST00000360909.3
ENST00000351081.1 ENST00000556135.1 |
DAAM1
|
dishevelled associated activator of morphogenesis 1 |
| chr1_+_4714792 | 0.02 |
ENST00000378190.3
|
AJAP1
|
adherens junctions associated protein 1 |
| chr14_+_39644425 | 0.02 |
ENST00000556530.1
|
PNN
|
pinin, desmosome associated protein |
| chr12_-_57882498 | 0.02 |
ENST00000550288.1
|
ARHGAP9
|
Rho GTPase activating protein 9 |
| chr14_+_24584372 | 0.02 |
ENST00000559396.1
ENST00000558638.1 ENST00000561041.1 ENST00000559288.1 ENST00000558408.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr9_-_74525658 | 0.02 |
ENST00000333421.6
|
ABHD17B
|
abhydrolase domain containing 17B |
| chr16_-_19896220 | 0.02 |
ENST00000562469.1
ENST00000300571.2 |
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B |
| chr2_+_56411131 | 0.02 |
ENST00000407595.2
|
CCDC85A
|
coiled-coil domain containing 85A |
| chr12_-_52867569 | 0.02 |
ENST00000252250.6
|
KRT6C
|
keratin 6C |
| chr1_-_43205811 | 0.02 |
ENST00000372539.3
ENST00000296387.1 ENST00000539749.1 |
CLDN19
|
claudin 19 |
| chr6_-_122792919 | 0.02 |
ENST00000339697.4
|
SERINC1
|
serine incorporator 1 |
| chr16_+_89724188 | 0.02 |
ENST00000301031.4
ENST00000566204.1 ENST00000579310.1 |
SPATA33
|
spermatogenesis associated 33 |
| chr2_-_37311445 | 0.02 |
ENST00000233099.5
ENST00000354531.2 |
HEATR5B
|
HEAT repeat containing 5B |
| chr5_-_145214848 | 0.02 |
ENST00000505416.1
ENST00000334744.4 ENST00000358004.2 ENST00000511435.1 |
PRELID2
|
PRELI domain containing 2 |
| chr4_-_146101304 | 0.02 |
ENST00000447906.2
|
OTUD4
|
OTU domain containing 4 |
| chrX_-_109561294 | 0.02 |
ENST00000372059.2
ENST00000262844.5 |
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr8_+_22298578 | 0.02 |
ENST00000240139.5
ENST00000289963.8 ENST00000397775.3 |
PPP3CC
|
protein phosphatase 3, catalytic subunit, gamma isozyme |
| chr5_-_132113063 | 0.02 |
ENST00000378719.2
|
SEPT8
|
septin 8 |
| chr2_+_128180842 | 0.02 |
ENST00000402125.2
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr3_+_111578640 | 0.02 |
ENST00000393925.3
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr8_-_101964265 | 0.02 |
ENST00000395958.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr11_-_12030681 | 0.02 |
ENST00000529338.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr2_+_37311645 | 0.02 |
ENST00000281932.5
|
GPATCH11
|
G patch domain containing 11 |
| chr10_+_101419187 | 0.02 |
ENST00000370489.4
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr17_+_43972010 | 0.02 |
ENST00000334239.8
ENST00000446361.3 |
MAPT
|
microtubule-associated protein tau |
| chr5_-_132113083 | 0.02 |
ENST00000296873.7
|
SEPT8
|
septin 8 |
| chr8_+_9413410 | 0.02 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr7_-_150780487 | 0.02 |
ENST00000482202.1
|
TMUB1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr8_-_134584152 | 0.02 |
ENST00000521180.1
ENST00000517668.1 ENST00000319914.5 |
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr1_+_167906056 | 0.02 |
ENST00000367840.3
|
DCAF6
|
DDB1 and CUL4 associated factor 6 |
| chrX_-_151143140 | 0.02 |
ENST00000393914.3
ENST00000370328.3 ENST00000370325.1 |
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon |
| chr14_+_24584056 | 0.02 |
ENST00000561001.1
|
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr5_+_178286925 | 0.02 |
ENST00000322434.3
|
ZNF354B
|
zinc finger protein 354B |
| chr14_-_51411194 | 0.02 |
ENST00000544180.2
|
PYGL
|
phosphorylase, glycogen, liver |
| chr2_-_20550416 | 0.02 |
ENST00000403432.1
ENST00000424110.1 |
PUM2
|
pumilio RNA-binding family member 2 |
| chr12_-_52887034 | 0.02 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr8_-_74884399 | 0.02 |
ENST00000520210.1
ENST00000602840.1 |
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr17_-_58469329 | 0.02 |
ENST00000393003.3
|
USP32
|
ubiquitin specific peptidase 32 |
| chr19_-_45873642 | 0.02 |
ENST00000485403.2
ENST00000586856.1 ENST00000586131.1 ENST00000391940.4 ENST00000221481.6 ENST00000391944.3 ENST00000391945.4 |
ERCC2
|
excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| chr6_+_139456226 | 0.02 |
ENST00000367658.2
|
HECA
|
headcase homolog (Drosophila) |
| chr8_-_74884459 | 0.02 |
ENST00000522337.1
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr3_-_185542817 | 0.02 |
ENST00000382199.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr3_-_33759699 | 0.01 |
ENST00000399362.4
ENST00000359576.5 ENST00000307312.7 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr3_-_108308241 | 0.01 |
ENST00000295746.8
|
KIAA1524
|
KIAA1524 |
| chr12_-_56122426 | 0.01 |
ENST00000551173.1
|
CD63
|
CD63 molecule |
| chr7_+_30174574 | 0.01 |
ENST00000409688.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr22_+_38035459 | 0.01 |
ENST00000357436.4
|
SH3BP1
|
SH3-domain binding protein 1 |
| chr8_+_17354617 | 0.01 |
ENST00000470360.1
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr11_+_67056867 | 0.01 |
ENST00000514166.1
|
ANKRD13D
|
ankyrin repeat domain 13 family, member D |
| chr8_-_38853990 | 0.01 |
ENST00000456845.2
ENST00000397070.2 ENST00000517872.1 ENST00000412303.1 ENST00000456397.2 |
TM2D2
|
TM2 domain containing 2 |
| chr9_+_74764278 | 0.01 |
ENST00000238018.4
ENST00000376989.3 |
GDA
|
guanine deaminase |
| chrX_-_128977781 | 0.01 |
ENST00000357166.6
|
ZDHHC9
|
zinc finger, DHHC-type containing 9 |
| chr11_-_117747607 | 0.01 |
ENST00000540359.1
ENST00000539526.1 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr5_+_140071011 | 0.01 |
ENST00000230771.3
ENST00000509299.1 ENST00000503873.1 ENST00000435019.2 ENST00000437649.2 ENST00000432671.2 |
HARS2
|
histidyl-tRNA synthetase 2, mitochondrial |
| chr2_-_74730087 | 0.01 |
ENST00000341396.2
|
LBX2
|
ladybird homeobox 2 |
| chr9_-_74525847 | 0.01 |
ENST00000377041.2
|
ABHD17B
|
abhydrolase domain containing 17B |
| chr3_+_111578027 | 0.01 |
ENST00000431670.2
ENST00000412622.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr14_-_22005018 | 0.01 |
ENST00000546363.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr17_-_5372271 | 0.01 |
ENST00000225296.3
|
DHX33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr2_-_73460334 | 0.01 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chr11_-_117747327 | 0.01 |
ENST00000584230.1
ENST00000527429.1 ENST00000584394.1 ENST00000532984.1 |
FXYD6
FXYD6-FXYD2
|
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB7A_ZBTB7C
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1903461 | Okazaki fragment processing involved in mitotic DNA replication(GO:1903461) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.1 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.2 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.0 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.0 | 0.0 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.0 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.2 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.0 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.0 | 0.0 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.0 | 0.1 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.0 | GO:0000406 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.0 | GO:0008892 | guanine deaminase activity(GO:0008892) |