Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
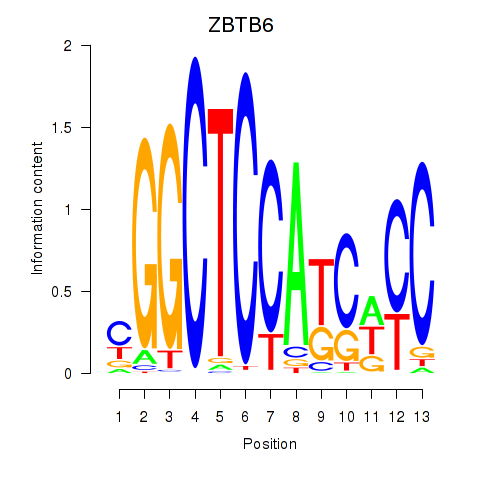
Results for ZBTB6
Z-value: 1.29
Transcription factors associated with ZBTB6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB6
|
ENSG00000186130.4 | zinc finger and BTB domain containing 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB6 | hg19_v2_chr9_-_125675576_125675612 | -0.51 | 3.0e-01 | Click! |
Activity profile of ZBTB6 motif
Sorted Z-values of ZBTB6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_66461175 | 1.08 |
ENST00000536005.2
ENST00000299694.8 ENST00000561796.1 |
BEAN1
|
brain expressed, associated with NEDD4, 1 |
| chr3_-_133614297 | 0.73 |
ENST00000486858.1
ENST00000477759.1 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chr17_-_43339474 | 0.65 |
ENST00000331780.4
|
SPATA32
|
spermatogenesis associated 32 |
| chr11_+_65686952 | 0.63 |
ENST00000527119.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr19_+_18530146 | 0.60 |
ENST00000348495.6
ENST00000270061.7 |
SSBP4
|
single stranded DNA binding protein 4 |
| chr4_+_159131596 | 0.56 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr16_-_67450325 | 0.56 |
ENST00000348579.2
|
ZDHHC1
|
zinc finger, DHHC-type containing 1 |
| chr17_+_40688190 | 0.56 |
ENST00000225927.2
|
NAGLU
|
N-acetylglucosaminidase, alpha |
| chr16_-_28503357 | 0.54 |
ENST00000333496.9
ENST00000561505.1 ENST00000567963.1 ENST00000354630.5 ENST00000355477.5 ENST00000357076.5 ENST00000565688.1 ENST00000359984.7 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr16_-_28503080 | 0.53 |
ENST00000565316.1
ENST00000565778.1 ENST00000357857.9 ENST00000568558.1 ENST00000357806.7 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr10_+_127585118 | 0.50 |
ENST00000449042.2
|
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr11_+_65383227 | 0.49 |
ENST00000355703.3
|
PCNXL3
|
pecanex-like 3 (Drosophila) |
| chr2_+_27282134 | 0.49 |
ENST00000441931.1
|
AGBL5
|
ATP/GTP binding protein-like 5 |
| chr17_-_43339453 | 0.47 |
ENST00000543122.1
|
SPATA32
|
spermatogenesis associated 32 |
| chr1_+_2160134 | 0.43 |
ENST00000378536.4
|
SKI
|
v-ski avian sarcoma viral oncogene homolog |
| chr10_+_81892477 | 0.43 |
ENST00000372263.3
|
PLAC9
|
placenta-specific 9 |
| chr8_-_72756667 | 0.43 |
ENST00000325509.4
|
MSC
|
musculin |
| chr11_-_615570 | 0.42 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr19_+_17830051 | 0.41 |
ENST00000594625.1
ENST00000324096.4 ENST00000600186.1 ENST00000597735.1 |
MAP1S
|
microtubule-associated protein 1S |
| chr11_+_65687158 | 0.40 |
ENST00000532933.1
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr19_-_18391708 | 0.40 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chr1_-_16532985 | 0.39 |
ENST00000441785.1
ENST00000449495.1 |
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr8_+_144451039 | 0.39 |
ENST00000289013.6
|
RHPN1
|
rhophilin, Rho GTPase binding protein 1 |
| chr11_+_82868185 | 0.39 |
ENST00000530304.1
ENST00000533018.1 |
PCF11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr1_+_43996518 | 0.39 |
ENST00000359947.4
ENST00000438120.1 |
PTPRF
|
protein tyrosine phosphatase, receptor type, F |
| chr16_-_28503327 | 0.38 |
ENST00000535392.1
ENST00000395653.4 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr22_+_39868786 | 0.38 |
ENST00000429402.1
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
| chr12_-_1703331 | 0.37 |
ENST00000339235.3
|
FBXL14
|
F-box and leucine-rich repeat protein 14 |
| chr17_+_54671047 | 0.37 |
ENST00000332822.4
|
NOG
|
noggin |
| chr22_+_38004723 | 0.37 |
ENST00000381756.5
|
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr19_+_34972543 | 0.37 |
ENST00000590071.2
|
WTIP
|
Wilms tumor 1 interacting protein |
| chr11_-_615942 | 0.37 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr9_-_116062045 | 0.36 |
ENST00000478815.1
|
RNF183
|
ring finger protein 183 |
| chr22_-_38484922 | 0.36 |
ENST00000428572.1
|
BAIAP2L2
|
BAI1-associated protein 2-like 2 |
| chrX_-_152736013 | 0.36 |
ENST00000330912.2
ENST00000338525.2 ENST00000334497.2 ENST00000370232.1 ENST00000370212.3 ENST00000370211.4 |
TREX2
HAUS7
|
three prime repair exonuclease 2 HAUS augmin-like complex, subunit 7 |
| chr14_-_69262916 | 0.36 |
ENST00000553375.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr19_-_4902877 | 0.35 |
ENST00000381781.2
|
ARRDC5
|
arrestin domain containing 5 |
| chr16_+_67197288 | 0.35 |
ENST00000264009.8
ENST00000421453.1 |
HSF4
|
heat shock transcription factor 4 |
| chr15_-_74504597 | 0.35 |
ENST00000416286.3
|
STRA6
|
stimulated by retinoic acid 6 |
| chr22_-_19419205 | 0.35 |
ENST00000340170.4
ENST00000263208.5 |
HIRA
|
histone cell cycle regulator |
| chr16_-_4664382 | 0.35 |
ENST00000591113.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr5_+_92919043 | 0.35 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr14_+_105266933 | 0.35 |
ENST00000555360.1
|
ZBTB42
|
zinc finger and BTB domain containing 42 |
| chr20_-_48532046 | 0.34 |
ENST00000543716.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr20_-_48532019 | 0.34 |
ENST00000289431.5
|
SPATA2
|
spermatogenesis associated 2 |
| chr6_-_30712313 | 0.34 |
ENST00000376377.2
ENST00000259874.5 |
IER3
|
immediate early response 3 |
| chr19_-_18717627 | 0.34 |
ENST00000392386.3
|
CRLF1
|
cytokine receptor-like factor 1 |
| chr16_+_30751953 | 0.34 |
ENST00000483578.1
|
RP11-2C24.4
|
RP11-2C24.4 |
| chr11_-_798305 | 0.33 |
ENST00000531514.1
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr9_-_123691047 | 0.33 |
ENST00000373887.3
|
TRAF1
|
TNF receptor-associated factor 1 |
| chr17_-_7232585 | 0.33 |
ENST00000571887.1
ENST00000315614.7 ENST00000399464.2 ENST00000570460.1 |
NEURL4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr11_-_66313699 | 0.33 |
ENST00000526986.1
ENST00000310442.3 |
ZDHHC24
|
zinc finger, DHHC-type containing 24 |
| chr19_+_54058073 | 0.33 |
ENST00000505949.1
ENST00000513265.1 |
ZNF331
|
zinc finger protein 331 |
| chr16_+_2039946 | 0.33 |
ENST00000248121.2
ENST00000568896.1 |
SYNGR3
|
synaptogyrin 3 |
| chr8_-_80942061 | 0.33 |
ENST00000519386.1
|
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr17_-_45918539 | 0.32 |
ENST00000584123.1
ENST00000578323.1 ENST00000407215.3 ENST00000290216.9 |
SCRN2
|
secernin 2 |
| chr15_-_75199213 | 0.32 |
ENST00000562698.1
|
FAM219B
|
family with sequence similarity 219, member B |
| chr16_+_1756162 | 0.32 |
ENST00000250894.4
ENST00000356010.5 |
MAPK8IP3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr8_-_19614810 | 0.31 |
ENST00000524213.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr15_-_74504560 | 0.31 |
ENST00000449139.2
|
STRA6
|
stimulated by retinoic acid 6 |
| chr8_-_144691718 | 0.31 |
ENST00000377579.3
ENST00000433751.1 ENST00000220966.6 |
PYCRL
|
pyrroline-5-carboxylate reductase-like |
| chr22_-_46283597 | 0.31 |
ENST00000451118.1
|
WI2-85898F10.1
|
WI2-85898F10.1 |
| chr15_-_43559055 | 0.31 |
ENST00000220420.5
ENST00000349114.4 |
TGM5
|
transglutaminase 5 |
| chr16_-_30621663 | 0.31 |
ENST00000287461.3
|
ZNF689
|
zinc finger protein 689 |
| chr22_+_31489344 | 0.31 |
ENST00000404574.1
|
SMTN
|
smoothelin |
| chr8_-_57123815 | 0.30 |
ENST00000316981.3
ENST00000423799.2 ENST00000429357.2 |
PLAG1
|
pleiomorphic adenoma gene 1 |
| chr22_+_38005033 | 0.30 |
ENST00000447515.1
ENST00000406772.1 ENST00000431745.1 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr11_-_1606513 | 0.30 |
ENST00000382171.2
|
KRTAP5-1
|
keratin associated protein 5-1 |
| chr20_-_62129163 | 0.30 |
ENST00000298049.7
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr8_-_145159083 | 0.30 |
ENST00000398712.2
|
SHARPIN
|
SHANK-associated RH domain interactor |
| chr3_-_129035120 | 0.30 |
ENST00000333762.4
|
H1FX
|
H1 histone family, member X |
| chr10_-_46342675 | 0.29 |
ENST00000492347.1
|
AGAP4
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 4 |
| chr5_-_149682447 | 0.29 |
ENST00000328668.7
|
ARSI
|
arylsulfatase family, member I |
| chr11_+_826136 | 0.29 |
ENST00000528315.1
ENST00000533803.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr22_-_20104700 | 0.29 |
ENST00000439169.2
ENST00000445045.1 ENST00000404751.3 ENST00000252136.7 ENST00000403707.3 |
TRMT2A
|
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
| chr22_+_38004832 | 0.29 |
ENST00000405147.3
ENST00000429218.1 ENST00000325180.8 ENST00000337437.4 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr2_+_220492373 | 0.29 |
ENST00000317151.3
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr22_-_31536480 | 0.29 |
ENST00000215885.3
|
PLA2G3
|
phospholipase A2, group III |
| chr19_-_55677999 | 0.29 |
ENST00000532817.1
ENST00000527223.2 ENST00000391720.4 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr16_-_2827128 | 0.28 |
ENST00000494946.2
ENST00000409477.1 ENST00000572954.1 ENST00000262306.7 ENST00000409906.4 |
TCEB2
|
transcription elongation factor B (SIII), polypeptide 2 (18kDa, elongin B) |
| chr16_-_4039001 | 0.28 |
ENST00000576936.1
|
ADCY9
|
adenylate cyclase 9 |
| chr19_-_55672037 | 0.28 |
ENST00000588076.1
|
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr8_-_109799793 | 0.28 |
ENST00000297459.3
|
TMEM74
|
transmembrane protein 74 |
| chr9_-_139581848 | 0.28 |
ENST00000538402.1
ENST00000371694.3 |
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 |
| chr15_-_74501360 | 0.28 |
ENST00000323940.5
|
STRA6
|
stimulated by retinoic acid 6 |
| chr11_+_65686728 | 0.28 |
ENST00000312515.2
ENST00000525501.1 |
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr1_-_26633480 | 0.27 |
ENST00000450041.1
|
UBXN11
|
UBX domain protein 11 |
| chr16_-_30905584 | 0.27 |
ENST00000380317.4
|
BCL7C
|
B-cell CLL/lymphoma 7C |
| chr20_+_2633269 | 0.27 |
ENST00000445139.1
|
NOP56
|
NOP56 ribonucleoprotein |
| chr17_+_1633755 | 0.27 |
ENST00000545662.1
|
WDR81
|
WD repeat domain 81 |
| chr3_-_48470838 | 0.27 |
ENST00000358459.4
ENST00000358536.4 |
PLXNB1
|
plexin B1 |
| chr20_+_34287194 | 0.27 |
ENST00000374078.1
ENST00000374077.3 |
ROMO1
|
reactive oxygen species modulator 1 |
| chr15_-_74045088 | 0.27 |
ENST00000569673.1
|
C15orf59
|
chromosome 15 open reading frame 59 |
| chr19_+_41698927 | 0.26 |
ENST00000310054.4
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr17_+_40119801 | 0.26 |
ENST00000585452.1
|
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr17_+_40118805 | 0.26 |
ENST00000591072.1
ENST00000587679.1 ENST00000393888.1 ENST00000441615.2 |
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr3_+_157261116 | 0.26 |
ENST00000468043.1
ENST00000459838.1 ENST00000461040.1 ENST00000449199.2 ENST00000426338.2 |
C3orf55
|
chromosome 3 open reading frame 55 |
| chr17_-_42402138 | 0.26 |
ENST00000592857.1
ENST00000586016.1 ENST00000590194.1 ENST00000377095.5 ENST00000588049.1 ENST00000586633.1 ENST00000537904.2 ENST00000585636.1 ENST00000585523.1 ENST00000225308.8 |
SLC25A39
|
solute carrier family 25, member 39 |
| chr20_+_34204939 | 0.26 |
ENST00000454819.1
|
SPAG4
|
sperm associated antigen 4 |
| chr4_-_140477910 | 0.26 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr19_+_54641444 | 0.26 |
ENST00000221232.5
ENST00000358389.3 |
CNOT3
|
CCR4-NOT transcription complex, subunit 3 |
| chr11_-_65359947 | 0.26 |
ENST00000597463.1
|
AP001362.1
|
Uncharacterized protein |
| chr11_+_45944190 | 0.26 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr9_-_139581875 | 0.26 |
ENST00000371696.2
|
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 |
| chr2_+_220492116 | 0.26 |
ENST00000373760.2
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr2_+_220492287 | 0.26 |
ENST00000273063.6
ENST00000373762.3 |
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr17_-_1553346 | 0.26 |
ENST00000301336.6
|
RILP
|
Rab interacting lysosomal protein |
| chr9_+_140149625 | 0.26 |
ENST00000343053.4
|
NELFB
|
negative elongation factor complex member B |
| chr10_+_88718397 | 0.26 |
ENST00000372017.3
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1) |
| chr22_+_38004473 | 0.25 |
ENST00000414350.3
ENST00000343632.4 |
GGA1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr22_+_24820341 | 0.25 |
ENST00000464977.1
ENST00000444262.2 |
ADORA2A
|
adenosine A2a receptor |
| chr1_-_26633067 | 0.25 |
ENST00000421827.2
ENST00000374215.1 ENST00000374223.1 ENST00000357089.4 ENST00000535108.1 ENST00000314675.7 ENST00000436301.2 ENST00000423664.1 ENST00000374221.3 |
UBXN11
|
UBX domain protein 11 |
| chr17_+_80416482 | 0.24 |
ENST00000309794.11
ENST00000345415.7 ENST00000457415.3 ENST00000584411.1 ENST00000412079.2 ENST00000577432.1 |
NARF
|
nuclear prelamin A recognition factor |
| chr8_+_99956662 | 0.24 |
ENST00000523368.1
ENST00000297565.4 ENST00000435298.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr8_-_80942139 | 0.24 |
ENST00000521434.1
ENST00000519120.1 ENST00000520946.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr6_+_157099036 | 0.24 |
ENST00000350026.5
ENST00000346085.5 ENST00000367148.1 ENST00000275248.4 |
ARID1B
|
AT rich interactive domain 1B (SWI1-like) |
| chr19_+_39926791 | 0.24 |
ENST00000594990.1
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr8_+_63161550 | 0.24 |
ENST00000328472.5
|
NKAIN3
|
Na+/K+ transporting ATPase interacting 3 |
| chr17_-_19265855 | 0.24 |
ENST00000440841.1
ENST00000395615.1 ENST00000461069.2 |
B9D1
|
B9 protein domain 1 |
| chr16_-_24506492 | 0.24 |
ENST00000566929.1
|
CTD-2313J23.1
|
CTD-2313J23.1 |
| chr1_-_223537401 | 0.24 |
ENST00000343846.3
ENST00000454695.2 ENST00000484758.2 |
SUSD4
|
sushi domain containing 4 |
| chr22_-_46933067 | 0.24 |
ENST00000262738.3
ENST00000395964.1 |
CELSR1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr8_-_80942467 | 0.24 |
ENST00000518271.1
ENST00000276585.4 ENST00000521605.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr15_+_85144217 | 0.24 |
ENST00000540936.1
ENST00000448803.2 ENST00000546275.1 ENST00000546148.1 ENST00000442073.3 ENST00000334141.3 ENST00000358472.3 ENST00000502939.2 ENST00000379358.3 ENST00000327179.6 |
ZSCAN2
|
zinc finger and SCAN domain containing 2 |
| chr12_-_50222187 | 0.24 |
ENST00000335999.6
|
NCKAP5L
|
NCK-associated protein 5-like |
| chr17_-_7518145 | 0.24 |
ENST00000250113.7
ENST00000571597.1 |
FXR2
|
fragile X mental retardation, autosomal homolog 2 |
| chr2_-_27603582 | 0.24 |
ENST00000323703.6
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chr6_+_29691056 | 0.24 |
ENST00000414333.1
ENST00000334668.4 ENST00000259951.7 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr2_+_74229812 | 0.24 |
ENST00000305799.7
|
TET3
|
tet methylcytosine dioxygenase 3 |
| chr17_-_80056099 | 0.23 |
ENST00000306749.2
|
FASN
|
fatty acid synthase |
| chr17_-_48227877 | 0.23 |
ENST00000316878.6
|
PPP1R9B
|
protein phosphatase 1, regulatory subunit 9B |
| chr1_+_26856236 | 0.23 |
ENST00000374168.2
ENST00000374166.4 |
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr8_-_145752390 | 0.23 |
ENST00000529415.2
ENST00000533758.1 |
LRRC24
|
leucine rich repeat containing 24 |
| chr1_+_152957707 | 0.23 |
ENST00000368762.1
|
SPRR1A
|
small proline-rich protein 1A |
| chr5_+_78985673 | 0.23 |
ENST00000446378.2
|
CMYA5
|
cardiomyopathy associated 5 |
| chr19_+_18530184 | 0.23 |
ENST00000601357.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chrX_-_153200676 | 0.23 |
ENST00000464845.1
|
NAA10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr2_+_220491973 | 0.23 |
ENST00000358055.3
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr19_+_57999079 | 0.22 |
ENST00000426954.2
ENST00000354197.4 ENST00000523882.1 ENST00000520540.1 ENST00000519310.1 ENST00000442920.2 ENST00000523312.1 ENST00000424930.2 |
ZNF419
|
zinc finger protein 419 |
| chr15_+_91411810 | 0.22 |
ENST00000268171.3
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr21_+_36041688 | 0.22 |
ENST00000360731.3
ENST00000349499.2 |
CLIC6
|
chloride intracellular channel 6 |
| chr19_+_41699135 | 0.22 |
ENST00000542619.1
ENST00000600561.1 |
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr10_+_103825080 | 0.22 |
ENST00000299238.5
|
HPS6
|
Hermansky-Pudlak syndrome 6 |
| chr22_+_43506747 | 0.22 |
ENST00000216115.2
|
BIK
|
BCL2-interacting killer (apoptosis-inducing) |
| chr9_+_136223414 | 0.22 |
ENST00000371964.4
|
SURF2
|
surfeit 2 |
| chr19_-_55770311 | 0.22 |
ENST00000412770.2
|
PPP6R1
|
protein phosphatase 6, regulatory subunit 1 |
| chr8_-_38325219 | 0.22 |
ENST00000533668.1
ENST00000413133.2 ENST00000397108.4 ENST00000526742.1 ENST00000525001.1 ENST00000425967.3 ENST00000529552.1 ENST00000397113.2 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr12_+_7037461 | 0.22 |
ENST00000396684.2
|
ATN1
|
atrophin 1 |
| chr3_+_10857885 | 0.22 |
ENST00000254488.2
ENST00000454147.1 |
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chr19_+_59055814 | 0.21 |
ENST00000594806.1
ENST00000253024.5 ENST00000341753.6 |
TRIM28
|
tripartite motif containing 28 |
| chr19_+_1495362 | 0.21 |
ENST00000395479.4
|
REEP6
|
receptor accessory protein 6 |
| chr22_+_38071615 | 0.21 |
ENST00000215909.5
|
LGALS1
|
lectin, galactoside-binding, soluble, 1 |
| chr8_-_146017736 | 0.21 |
ENST00000528957.1
|
RPL8
|
ribosomal protein L8 |
| chr1_+_43148059 | 0.21 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr9_+_135906076 | 0.21 |
ENST00000372097.5
ENST00000440319.1 |
GTF3C5
|
general transcription factor IIIC, polypeptide 5, 63kDa |
| chr12_-_92539614 | 0.21 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr20_+_42295745 | 0.21 |
ENST00000396863.4
ENST00000217026.4 |
MYBL2
|
v-myb avian myeloblastosis viral oncogene homolog-like 2 |
| chr22_+_45098067 | 0.21 |
ENST00000336985.6
ENST00000403696.1 ENST00000457960.1 ENST00000361473.5 |
PRR5
PRR5-ARHGAP8
|
proline rich 5 (renal) PRR5-ARHGAP8 readthrough |
| chr19_-_51529849 | 0.21 |
ENST00000600362.1
ENST00000453757.3 ENST00000601671.1 |
KLK11
|
kallikrein-related peptidase 11 |
| chr1_+_15272271 | 0.21 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chr20_+_34287364 | 0.21 |
ENST00000374072.1
ENST00000397416.1 ENST00000336695.4 |
ROMO1
|
reactive oxygen species modulator 1 |
| chr7_-_5463175 | 0.21 |
ENST00000399537.4
ENST00000430969.1 |
TNRC18
|
trinucleotide repeat containing 18 |
| chr19_+_57999101 | 0.21 |
ENST00000347466.6
ENST00000523138.1 ENST00000415379.2 ENST00000521754.1 ENST00000221735.7 ENST00000518999.1 ENST00000521137.1 |
ZNF419
|
zinc finger protein 419 |
| chr19_-_55677920 | 0.21 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr20_+_44509857 | 0.21 |
ENST00000372523.1
ENST00000372520.1 |
ZSWIM1
|
zinc finger, SWIM-type containing 1 |
| chr20_-_34287259 | 0.20 |
ENST00000397425.1
ENST00000540053.1 ENST00000541387.1 ENST00000374092.4 |
NFS1
|
NFS1 cysteine desulfurase |
| chr14_-_21994525 | 0.20 |
ENST00000538754.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr16_-_66968265 | 0.20 |
ENST00000567511.1
ENST00000422424.2 |
FAM96B
|
family with sequence similarity 96, member B |
| chr22_-_32058166 | 0.20 |
ENST00000435900.1
ENST00000336566.4 |
PISD
|
phosphatidylserine decarboxylase |
| chr7_+_102004322 | 0.20 |
ENST00000496391.1
|
PRKRIP1
|
PRKR interacting protein 1 (IL11 inducible) |
| chr16_+_57680043 | 0.20 |
ENST00000569154.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr17_+_80416050 | 0.20 |
ENST00000579198.1
ENST00000390006.4 ENST00000580296.1 |
NARF
|
nuclear prelamin A recognition factor |
| chrY_-_15591485 | 0.20 |
ENST00000382896.4
ENST00000537580.1 ENST00000540140.1 ENST00000545955.1 ENST00000538878.1 |
UTY
|
ubiquitously transcribed tetratricopeptide repeat containing, Y-linked |
| chr22_-_31741757 | 0.19 |
ENST00000215919.3
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr22_-_50216853 | 0.19 |
ENST00000342989.5
ENST00000542442.1 |
BRD1
|
bromodomain containing 1 |
| chr16_-_31021921 | 0.19 |
ENST00000215095.5
|
STX1B
|
syntaxin 1B |
| chr3_+_157261035 | 0.19 |
ENST00000312275.5
|
C3orf55
|
chromosome 3 open reading frame 55 |
| chr22_-_32058416 | 0.19 |
ENST00000439502.2
|
PISD
|
phosphatidylserine decarboxylase |
| chr2_-_27357479 | 0.19 |
ENST00000406567.3
ENST00000260643.2 |
PREB
|
prolactin regulatory element binding |
| chr3_-_133614467 | 0.19 |
ENST00000469959.1
|
RAB6B
|
RAB6B, member RAS oncogene family |
| chr6_-_72130472 | 0.19 |
ENST00000426635.2
|
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr8_-_145115584 | 0.19 |
ENST00000426825.1
|
OPLAH
|
5-oxoprolinase (ATP-hydrolysing) |
| chr11_-_62359027 | 0.19 |
ENST00000494385.1
ENST00000308436.7 |
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr2_+_108145913 | 0.18 |
ENST00000443205.1
|
AC096669.3
|
AC096669.3 |
| chr14_-_50999190 | 0.18 |
ENST00000557390.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr22_+_22764088 | 0.18 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chr19_-_3971050 | 0.18 |
ENST00000545797.2
ENST00000596311.1 |
DAPK3
|
death-associated protein kinase 3 |
| chr1_+_101361782 | 0.18 |
ENST00000357650.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr11_+_63753883 | 0.18 |
ENST00000538426.1
ENST00000543004.1 |
OTUB1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr12_+_113659234 | 0.18 |
ENST00000551096.1
ENST00000551099.1 ENST00000335509.6 ENST00000552897.1 ENST00000550785.1 ENST00000549279.1 |
TPCN1
|
two pore segment channel 1 |
| chr1_-_153935791 | 0.18 |
ENST00000429040.1
|
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr1_-_40782938 | 0.18 |
ENST00000372736.3
ENST00000372748.3 |
COL9A2
|
collagen, type IX, alpha 2 |
| chrX_-_153200411 | 0.18 |
ENST00000393712.3
ENST00000370009.1 ENST00000370011.3 ENST00000370015.4 |
NAA10
|
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr17_-_41910505 | 0.18 |
ENST00000398389.4
|
MPP3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) |
| chr17_+_15603447 | 0.18 |
ENST00000395893.2
|
ZNF286A
|
Homo sapiens zinc finger protein 286A (ZNF286A), transcript variant 6, mRNA. |
| chr9_+_34646651 | 0.18 |
ENST00000378842.3
|
GALT
|
galactose-1-phosphate uridylyltransferase |
| chr12_+_132628963 | 0.18 |
ENST00000330579.1
|
NOC4L
|
nucleolar complex associated 4 homolog (S. cerevisiae) |
| chr5_-_178054014 | 0.18 |
ENST00000520957.1
|
CLK4
|
CDC-like kinase 4 |
| chr19_-_14201507 | 0.18 |
ENST00000533683.2
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr10_+_30723045 | 0.18 |
ENST00000542547.1
ENST00000415139.1 |
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.3 | 0.3 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.2 | 0.8 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.2 | 1.5 | GO:0043585 | nose morphogenesis(GO:0043585) |
| 0.2 | 0.5 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.6 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.1 | 0.4 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.1 | 0.3 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.1 | 0.4 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.4 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.3 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.2 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.1 | 0.3 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.2 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 | 0.2 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 0.2 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.1 | 0.2 | GO:0090472 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.1 | 0.4 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.1 | 0.2 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.1 | 0.3 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.3 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.1 | 0.2 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 0.3 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.1 | 0.4 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.1 | 0.2 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) glomerular endothelium development(GO:0072011) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.1 | 0.2 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.8 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.3 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.2 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.0 | 0.2 | GO:1901535 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.2 | GO:0072312 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.1 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.3 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.3 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.0 | 0.3 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.5 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.4 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.0 | 0.2 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.2 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.2 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.6 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.3 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.4 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.1 | GO:0043309 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) regulation of eosinophil activation(GO:1902566) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.6 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) |
| 0.0 | 0.3 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) histone H2A K63-linked ubiquitination(GO:0070535) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.3 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.2 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.2 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) negative regulation of mast cell cytokine production(GO:0032764) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.0 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.1 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.0 | 0.6 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.1 | GO:2000051 | Wnt receptor catabolic process(GO:0038018) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0032796 | uropod organization(GO:0032796) |
| 0.0 | 0.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.3 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.3 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.3 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 1.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.3 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.0 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 1.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.2 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.0 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.5 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.9 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.4 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.0 | GO:2000506 | negative regulation of energy homeostasis(GO:2000506) |
| 0.0 | 0.1 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.0 | GO:1904978 | regulation of endosome organization(GO:1904978) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.0 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.0 | 0.0 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.2 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0036284 | tubulobulbar complex(GO:0036284) |
| 0.1 | 0.5 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 0.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 0.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.6 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.2 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.1 | 0.2 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.3 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.5 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.0 | 0.3 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.3 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 1.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0097513 | actomyosin contractile ring(GO:0005826) myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.0 | GO:0039714 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.4 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.0 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.5 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.1 | 0.3 | GO:0001003 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.1 | 0.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.4 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 0.6 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 0.2 | GO:0031177 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.4 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 1.0 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.3 | GO:0052812 | phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 1.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.2 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.2 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0031071 | cysteine desulfurase activity(GO:0031071) |
| 0.0 | 0.3 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 1.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.3 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.2 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.3 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.3 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.1 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 0.3 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.2 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.7 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.6 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0070123 | transforming growth factor beta receptor activity, type III(GO:0070123) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.0 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.3 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.0 | 0.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.0 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.0 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.8 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.2 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |