Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
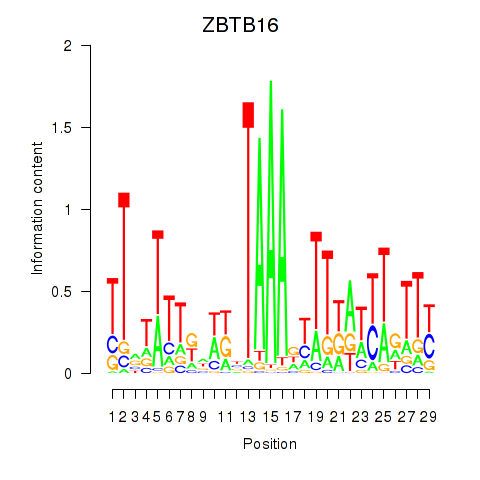
Results for ZBTB16
Z-value: 0.34
Transcription factors associated with ZBTB16
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB16
|
ENSG00000109906.9 | zinc finger and BTB domain containing 16 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB16 | hg19_v2_chr11_+_113930955_113930979 | 0.67 | 1.5e-01 | Click! |
Activity profile of ZBTB16 motif
Sorted Z-values of ZBTB16 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_74301880 | 0.36 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr10_-_98347063 | 0.24 |
ENST00000443638.1
|
TM9SF3
|
transmembrane 9 superfamily member 3 |
| chr12_-_53601000 | 0.21 |
ENST00000338737.4
ENST00000549086.2 |
ITGB7
|
integrin, beta 7 |
| chr12_-_10601963 | 0.21 |
ENST00000543893.1
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr1_-_100231349 | 0.20 |
ENST00000287474.5
ENST00000414213.1 |
FRRS1
|
ferric-chelate reductase 1 |
| chr2_-_188419078 | 0.20 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr2_+_181988620 | 0.17 |
ENST00000428474.1
ENST00000424655.1 |
AC104820.2
|
AC104820.2 |
| chr15_+_66585555 | 0.17 |
ENST00000319194.5
ENST00000525134.2 ENST00000441424.2 |
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr19_-_53324884 | 0.17 |
ENST00000457749.2
ENST00000414252.2 ENST00000391783.2 |
ZNF28
|
zinc finger protein 28 |
| chr8_-_48651648 | 0.17 |
ENST00000408965.3
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr12_-_53601055 | 0.16 |
ENST00000552972.1
ENST00000422257.3 ENST00000267082.5 |
ITGB7
|
integrin, beta 7 |
| chr14_+_67656110 | 0.16 |
ENST00000524532.1
ENST00000530728.1 |
FAM71D
|
family with sequence similarity 71, member D |
| chr12_+_97306295 | 0.16 |
ENST00000457368.2
|
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr5_-_24645078 | 0.15 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr16_-_14726576 | 0.15 |
ENST00000562896.1
|
PARN
|
poly(A)-specific ribonuclease |
| chr18_-_52626622 | 0.14 |
ENST00000591504.1
|
CCDC68
|
coiled-coil domain containing 68 |
| chr9_-_15472730 | 0.13 |
ENST00000481862.1
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr19_+_21264943 | 0.13 |
ENST00000597424.1
|
ZNF714
|
zinc finger protein 714 |
| chr5_+_178322893 | 0.13 |
ENST00000361362.2
ENST00000520660.1 ENST00000520805.1 |
ZFP2
|
ZFP2 zinc finger protein |
| chr19_-_45004556 | 0.13 |
ENST00000587047.1
ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180
|
zinc finger protein 180 |
| chr10_+_111765562 | 0.13 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr13_-_101240985 | 0.13 |
ENST00000471912.1
|
GGACT
|
gamma-glutamylamine cyclotransferase |
| chr10_+_104613980 | 0.12 |
ENST00000339834.5
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr10_+_62538089 | 0.12 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr3_-_155524049 | 0.12 |
ENST00000534941.1
ENST00000340171.2 |
C3orf33
|
chromosome 3 open reading frame 33 |
| chr18_+_6256746 | 0.12 |
ENST00000578427.1
|
RP11-760N9.1
|
RP11-760N9.1 |
| chr18_+_39535239 | 0.12 |
ENST00000585528.1
|
PIK3C3
|
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr8_+_67687773 | 0.12 |
ENST00000518388.1
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr5_+_180682720 | 0.12 |
ENST00000599439.1
|
AC008443.1
|
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
| chr3_-_156272924 | 0.12 |
ENST00000467789.1
ENST00000265044.2 |
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr19_-_23869999 | 0.12 |
ENST00000601935.1
ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675
|
zinc finger protein 675 |
| chr19_+_9296279 | 0.11 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr15_+_45028719 | 0.11 |
ENST00000560442.1
ENST00000558329.1 ENST00000561043.1 |
TRIM69
|
tripartite motif containing 69 |
| chr12_-_85430024 | 0.11 |
ENST00000547836.1
ENST00000532498.2 |
TSPAN19
|
tetraspanin 19 |
| chr19_+_21264980 | 0.11 |
ENST00000596053.1
ENST00000597086.1 ENST00000596143.1 ENST00000596367.1 ENST00000601416.1 |
ZNF714
|
zinc finger protein 714 |
| chr12_-_15865844 | 0.11 |
ENST00000543612.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr8_+_104831472 | 0.11 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr19_-_19932501 | 0.11 |
ENST00000540806.2
ENST00000590766.1 ENST00000587452.1 ENST00000545006.1 ENST00000590319.1 ENST00000587461.1 ENST00000450683.2 ENST00000443905.2 ENST00000590274.1 |
ZNF506
CTC-559E9.4
|
zinc finger protein 506 CTC-559E9.4 |
| chr7_-_7606626 | 0.11 |
ENST00000609497.1
|
RP5-1159O4.1
|
RP5-1159O4.1 |
| chr13_+_77564795 | 0.10 |
ENST00000377453.3
|
CLN5
|
ceroid-lipofuscinosis, neuronal 5 |
| chr3_-_93692781 | 0.10 |
ENST00000394236.3
|
PROS1
|
protein S (alpha) |
| chr5_+_68860949 | 0.10 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr10_+_134258649 | 0.10 |
ENST00000392630.3
ENST00000321248.2 |
C10orf91
|
chromosome 10 open reading frame 91 |
| chr18_+_11857439 | 0.10 |
ENST00000602628.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr3_+_183353356 | 0.10 |
ENST00000242810.6
ENST00000493074.1 ENST00000437402.1 ENST00000454495.2 ENST00000473045.1 ENST00000468101.1 ENST00000427201.2 ENST00000482138.1 ENST00000454652.2 |
KLHL24
|
kelch-like family member 24 |
| chr8_+_66619277 | 0.09 |
ENST00000521247.2
ENST00000527155.1 |
MTFR1
|
mitochondrial fission regulator 1 |
| chr5_+_67586465 | 0.09 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr8_-_30585439 | 0.09 |
ENST00000221130.5
|
GSR
|
glutathione reductase |
| chr3_-_138312971 | 0.09 |
ENST00000485115.1
ENST00000484888.1 ENST00000468900.1 ENST00000542237.1 ENST00000481834.1 |
CEP70
|
centrosomal protein 70kDa |
| chr18_-_21017817 | 0.09 |
ENST00000542162.1
ENST00000383233.3 ENST00000582336.1 ENST00000450466.2 ENST00000578520.1 ENST00000399707.1 |
TMEM241
|
transmembrane protein 241 |
| chr7_+_112120908 | 0.09 |
ENST00000439068.2
ENST00000312849.4 ENST00000429049.1 |
LSMEM1
|
leucine-rich single-pass membrane protein 1 |
| chr12_+_133757995 | 0.09 |
ENST00000536435.2
ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr19_-_46580352 | 0.09 |
ENST00000601672.1
|
IGFL4
|
IGF-like family member 4 |
| chr12_+_133758115 | 0.09 |
ENST00000541009.2
ENST00000592241.1 |
ZNF268
|
zinc finger protein 268 |
| chr3_-_138313161 | 0.09 |
ENST00000489254.1
ENST00000474781.1 ENST00000462419.1 ENST00000264982.3 |
CEP70
|
centrosomal protein 70kDa |
| chr15_-_52944231 | 0.09 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr4_+_95128748 | 0.09 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr9_-_110540419 | 0.09 |
ENST00000398726.3
|
AL162389.1
|
Uncharacterized protein |
| chr17_-_46623441 | 0.08 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr1_+_33439268 | 0.08 |
ENST00000594612.1
|
FKSG48
|
FKSG48 |
| chr7_+_120591170 | 0.08 |
ENST00000431467.1
|
ING3
|
inhibitor of growth family, member 3 |
| chr22_+_29168652 | 0.08 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr16_-_20817753 | 0.08 |
ENST00000389345.5
ENST00000300005.3 ENST00000357967.4 ENST00000569729.1 |
ERI2
|
ERI1 exoribonuclease family member 2 |
| chr8_-_8318847 | 0.08 |
ENST00000521218.1
|
CTA-398F10.2
|
CTA-398F10.2 |
| chr3_-_93747425 | 0.08 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr7_-_130066571 | 0.08 |
ENST00000492389.1
|
CEP41
|
centrosomal protein 41kDa |
| chr1_+_76251912 | 0.08 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr13_+_76123883 | 0.08 |
ENST00000377595.3
|
UCHL3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr16_-_18887627 | 0.08 |
ENST00000563235.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr4_+_28437071 | 0.08 |
ENST00000509416.1
|
RP11-123O22.1
|
RP11-123O22.1 |
| chr2_-_11606275 | 0.07 |
ENST00000381525.3
ENST00000362009.4 |
E2F6
|
E2F transcription factor 6 |
| chr19_+_24269981 | 0.07 |
ENST00000339642.6
ENST00000357002.4 |
ZNF254
|
zinc finger protein 254 |
| chr3_-_165555200 | 0.07 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr4_-_185275104 | 0.07 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr11_+_120255997 | 0.07 |
ENST00000532993.1
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr12_-_25150409 | 0.07 |
ENST00000549262.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr1_+_204494618 | 0.07 |
ENST00000367180.1
ENST00000391947.2 |
MDM4
|
Mdm4 p53 binding protein homolog (mouse) |
| chr11_-_111649074 | 0.07 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr21_+_40817749 | 0.06 |
ENST00000380637.3
ENST00000380634.1 ENST00000458295.1 ENST00000440288.2 ENST00000380631.1 |
SH3BGR
|
SH3 domain binding glutamic acid-rich protein |
| chr16_-_70239683 | 0.06 |
ENST00000601706.1
|
AC009060.1
|
Uncharacterized protein |
| chr7_+_28725585 | 0.06 |
ENST00000396298.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr12_+_4385230 | 0.06 |
ENST00000536537.1
|
CCND2
|
cyclin D2 |
| chr15_+_69365272 | 0.06 |
ENST00000559914.1
ENST00000558369.1 |
LINC00277
|
long intergenic non-protein coding RNA 277 |
| chr21_-_30391636 | 0.06 |
ENST00000493196.1
|
RWDD2B
|
RWD domain containing 2B |
| chr4_-_99851766 | 0.06 |
ENST00000450253.2
|
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr14_+_52164820 | 0.06 |
ENST00000554167.1
|
FRMD6
|
FERM domain containing 6 |
| chr17_+_19314432 | 0.06 |
ENST00000575165.2
|
RNF112
|
ring finger protein 112 |
| chr17_+_46908350 | 0.06 |
ENST00000258947.3
ENST00000509507.1 ENST00000448105.2 ENST00000570513.1 ENST00000509415.1 ENST00000513119.1 ENST00000416445.2 ENST00000508679.1 ENST00000505071.1 |
CALCOCO2
|
calcium binding and coiled-coil domain 2 |
| chr15_-_55562451 | 0.06 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr20_-_33264886 | 0.06 |
ENST00000217446.3
ENST00000452740.2 ENST00000374820.2 |
PIGU
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr19_+_21265028 | 0.06 |
ENST00000291770.7
|
ZNF714
|
zinc finger protein 714 |
| chr12_+_85430110 | 0.06 |
ENST00000393212.3
ENST00000393217.2 |
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1 |
| chr5_-_54988448 | 0.06 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr3_+_184529948 | 0.05 |
ENST00000436792.2
ENST00000446204.2 ENST00000422105.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr3_+_179065474 | 0.05 |
ENST00000471841.1
ENST00000280653.7 |
MFN1
|
mitofusin 1 |
| chr1_+_104159999 | 0.05 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr21_-_40817645 | 0.05 |
ENST00000438404.1
ENST00000358268.2 ENST00000411566.1 ENST00000451131.1 ENST00000418018.1 ENST00000415863.1 ENST00000426783.1 ENST00000288350.3 ENST00000485895.2 ENST00000448288.2 ENST00000456017.1 ENST00000434281.1 |
LCA5L
|
Leber congenital amaurosis 5-like |
| chr4_-_99850243 | 0.05 |
ENST00000280892.6
ENST00000511644.1 ENST00000504432.1 ENST00000505992.1 |
EIF4E
|
eukaryotic translation initiation factor 4E |
| chrX_+_23925918 | 0.05 |
ENST00000379211.3
|
CXorf58
|
chromosome X open reading frame 58 |
| chr10_-_74283694 | 0.05 |
ENST00000398763.4
ENST00000418483.2 ENST00000489666.2 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr2_+_89901292 | 0.05 |
ENST00000448155.2
|
IGKV1D-39
|
immunoglobulin kappa variable 1D-39 |
| chr1_+_43613566 | 0.05 |
ENST00000409396.1
|
FAM183A
|
family with sequence similarity 183, member A |
| chrX_-_30885319 | 0.05 |
ENST00000378933.1
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr4_-_159094194 | 0.05 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr12_-_75905374 | 0.05 |
ENST00000438169.2
ENST00000229214.4 |
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr22_-_36236623 | 0.05 |
ENST00000405409.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr12_+_21207503 | 0.05 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr22_-_38239808 | 0.05 |
ENST00000406423.1
ENST00000424350.1 ENST00000458278.2 |
ANKRD54
|
ankyrin repeat domain 54 |
| chr1_+_210501589 | 0.05 |
ENST00000413764.2
ENST00000541565.1 |
HHAT
|
hedgehog acyltransferase |
| chr4_-_48014931 | 0.05 |
ENST00000420489.2
ENST00000504722.1 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr16_+_24549014 | 0.05 |
ENST00000564314.1
ENST00000567686.1 |
RBBP6
|
retinoblastoma binding protein 6 |
| chr3_+_184529929 | 0.05 |
ENST00000287546.4
ENST00000437079.3 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr9_+_79074068 | 0.05 |
ENST00000444201.2
ENST00000376730.4 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2 |
| chr6_+_106988986 | 0.04 |
ENST00000457437.1
ENST00000535438.1 |
AIM1
|
absent in melanoma 1 |
| chr11_-_66445219 | 0.04 |
ENST00000525754.1
ENST00000531969.1 ENST00000524637.1 ENST00000531036.2 ENST00000310046.4 |
RBM4B
|
RNA binding motif protein 4B |
| chr12_-_74686314 | 0.04 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr7_+_16828866 | 0.04 |
ENST00000597084.1
|
AC073333.1
|
Uncharacterized protein |
| chr10_+_43916052 | 0.04 |
ENST00000442526.2
|
RP11-517P14.2
|
RP11-517P14.2 |
| chr14_+_77228532 | 0.04 |
ENST00000167106.4
ENST00000554237.1 |
VASH1
|
vasohibin 1 |
| chr20_-_18477862 | 0.04 |
ENST00000337227.4
|
RBBP9
|
retinoblastoma binding protein 9 |
| chr9_+_131062367 | 0.04 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr8_+_67687413 | 0.04 |
ENST00000521960.1
ENST00000522398.1 ENST00000522629.1 ENST00000520976.1 ENST00000396596.1 |
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr11_+_43380459 | 0.04 |
ENST00000299240.6
ENST00000039989.4 |
TTC17
|
tetratricopeptide repeat domain 17 |
| chr2_+_128458514 | 0.04 |
ENST00000310981.4
|
SFT2D3
|
SFT2 domain containing 3 |
| chr1_+_198608146 | 0.04 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr4_+_106631966 | 0.04 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr5_+_72509751 | 0.04 |
ENST00000515556.1
ENST00000513379.1 ENST00000427584.2 |
RP11-60A8.1
|
RP11-60A8.1 |
| chr21_+_37692481 | 0.04 |
ENST00000400485.1
|
MORC3
|
MORC family CW-type zinc finger 3 |
| chr4_+_86699834 | 0.04 |
ENST00000395183.2
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr3_+_160394940 | 0.04 |
ENST00000320767.2
|
ARL14
|
ADP-ribosylation factor-like 14 |
| chr2_+_200472779 | 0.04 |
ENST00000427045.1
ENST00000419243.1 |
AC093590.1
|
AC093590.1 |
| chr17_+_74463650 | 0.03 |
ENST00000392492.3
|
AANAT
|
aralkylamine N-acetyltransferase |
| chr4_+_96012585 | 0.03 |
ENST00000502683.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr4_-_39033963 | 0.03 |
ENST00000381938.3
|
TMEM156
|
transmembrane protein 156 |
| chr16_+_81070792 | 0.03 |
ENST00000564241.1
ENST00000565237.1 |
ATMIN
|
ATM interactor |
| chr1_+_15256230 | 0.03 |
ENST00000376028.4
ENST00000400798.2 |
KAZN
|
kazrin, periplakin interacting protein |
| chr5_-_34919094 | 0.03 |
ENST00000341754.4
|
RAD1
|
RAD1 homolog (S. pombe) |
| chr4_-_68995571 | 0.03 |
ENST00000356291.2
|
TMPRSS11F
|
transmembrane protease, serine 11F |
| chr14_+_39703084 | 0.03 |
ENST00000553728.1
|
RP11-407N17.3
|
cTAGE family member 5 isoform 4 |
| chr3_-_137893721 | 0.03 |
ENST00000505015.2
ENST00000260803.4 |
DBR1
|
debranching RNA lariats 1 |
| chr14_-_45722360 | 0.03 |
ENST00000451174.1
|
MIS18BP1
|
MIS18 binding protein 1 |
| chr6_-_34639733 | 0.03 |
ENST00000374021.1
|
C6orf106
|
chromosome 6 open reading frame 106 |
| chr6_+_25963020 | 0.03 |
ENST00000357085.3
|
TRIM38
|
tripartite motif containing 38 |
| chr16_+_81812863 | 0.03 |
ENST00000359376.3
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| chr12_-_68696652 | 0.03 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chrX_+_118602363 | 0.03 |
ENST00000317881.8
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr8_-_74205851 | 0.03 |
ENST00000396467.1
|
RPL7
|
ribosomal protein L7 |
| chr12_+_32832134 | 0.03 |
ENST00000452533.2
|
DNM1L
|
dynamin 1-like |
| chr3_+_44840679 | 0.03 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr7_+_134464414 | 0.03 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr2_+_242289502 | 0.03 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr22_+_24198890 | 0.03 |
ENST00000345044.6
|
SLC2A11
|
solute carrier family 2 (facilitated glucose transporter), member 11 |
| chr11_-_104972158 | 0.03 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr6_+_42749759 | 0.03 |
ENST00000314073.5
|
GLTSCR1L
|
GLTSCR1-like |
| chr1_+_32608566 | 0.03 |
ENST00000545542.1
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr11_+_65657875 | 0.03 |
ENST00000312579.2
|
CCDC85B
|
coiled-coil domain containing 85B |
| chr3_+_57875738 | 0.03 |
ENST00000417128.1
ENST00000438794.1 |
SLMAP
|
sarcolemma associated protein |
| chr11_-_111649015 | 0.03 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr12_+_32832203 | 0.03 |
ENST00000553257.1
ENST00000549701.1 ENST00000358214.5 ENST00000266481.6 ENST00000551476.1 ENST00000550154.1 ENST00000547312.1 ENST00000414834.2 ENST00000381000.4 ENST00000548750.1 |
DNM1L
|
dynamin 1-like |
| chr7_-_32534850 | 0.03 |
ENST00000409952.3
ENST00000409909.3 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr2_+_191334212 | 0.03 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr4_+_26323764 | 0.03 |
ENST00000514730.1
ENST00000507574.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr5_+_96212185 | 0.03 |
ENST00000379904.4
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr1_+_76251879 | 0.03 |
ENST00000535300.1
ENST00000319942.3 |
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr5_+_159848854 | 0.03 |
ENST00000517480.1
ENST00000520452.1 ENST00000393964.1 |
PTTG1
|
pituitary tumor-transforming 1 |
| chr14_+_67831576 | 0.03 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr1_+_178310581 | 0.03 |
ENST00000462775.1
|
RASAL2
|
RAS protein activator like 2 |
| chr4_+_96012614 | 0.03 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr2_-_61245363 | 0.03 |
ENST00000316752.6
|
PUS10
|
pseudouridylate synthase 10 |
| chr3_+_130569592 | 0.03 |
ENST00000533801.2
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr4_-_166034029 | 0.02 |
ENST00000306480.6
|
TMEM192
|
transmembrane protein 192 |
| chr6_+_134758827 | 0.02 |
ENST00000431422.1
|
LINC01010
|
long intergenic non-protein coding RNA 1010 |
| chr5_+_179135246 | 0.02 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr3_-_133969673 | 0.02 |
ENST00000427044.2
|
RYK
|
receptor-like tyrosine kinase |
| chr16_+_89984287 | 0.02 |
ENST00000555147.1
|
MC1R
|
melanocortin 1 receptor (alpha melanocyte stimulating hormone receptor) |
| chr18_+_39535152 | 0.02 |
ENST00000262039.4
ENST00000398870.3 ENST00000586545.1 |
PIK3C3
|
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr19_+_45542295 | 0.02 |
ENST00000221455.3
ENST00000391953.4 ENST00000588936.1 |
CLASRP
|
CLK4-associating serine/arginine rich protein |
| chr5_+_140705777 | 0.02 |
ENST00000606901.1
ENST00000606674.1 |
AC005618.6
|
AC005618.6 |
| chr18_+_7946839 | 0.02 |
ENST00000578916.1
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chrX_-_73834449 | 0.02 |
ENST00000332687.6
ENST00000349225.2 |
RLIM
|
ring finger protein, LIM domain interacting |
| chr2_-_32490859 | 0.02 |
ENST00000404025.2
|
NLRC4
|
NLR family, CARD domain containing 4 |
| chr18_-_64271363 | 0.02 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr2_+_192543694 | 0.02 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr19_+_13135386 | 0.02 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr8_+_67039131 | 0.02 |
ENST00000315962.4
ENST00000353317.5 |
TRIM55
|
tripartite motif containing 55 |
| chr16_-_18908196 | 0.02 |
ENST00000565324.1
ENST00000561947.1 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr12_+_20968608 | 0.02 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr15_-_54051831 | 0.02 |
ENST00000557913.1
ENST00000360509.5 |
WDR72
|
WD repeat domain 72 |
| chr6_-_138893661 | 0.02 |
ENST00000427025.2
|
NHSL1
|
NHS-like 1 |
| chr7_-_16872932 | 0.02 |
ENST00000419572.2
ENST00000412973.1 |
AGR2
|
anterior gradient 2 |
| chr5_+_140560980 | 0.02 |
ENST00000361016.2
|
PCDHB16
|
protocadherin beta 16 |
| chr19_-_58662139 | 0.02 |
ENST00000598312.1
|
ZNF329
|
zinc finger protein 329 |
| chr7_+_7606497 | 0.02 |
ENST00000340080.4
ENST00000405785.1 ENST00000433635.1 |
MIOS
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr18_+_33709834 | 0.02 |
ENST00000358232.6
ENST00000351393.6 ENST00000442325.2 ENST00000423854.2 ENST00000350494.6 ENST00000542824.1 |
ELP2
|
elongator acetyltransferase complex subunit 2 |
| chr1_+_174769006 | 0.02 |
ENST00000489615.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr5_-_124084493 | 0.02 |
ENST00000509799.1
|
ZNF608
|
zinc finger protein 608 |
| chr12_+_26164645 | 0.02 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr1_-_115292591 | 0.02 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr11_-_70672645 | 0.02 |
ENST00000423696.2
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr4_-_90758118 | 0.02 |
ENST00000420646.2
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB16
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 0.2 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.4 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.1 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.1 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.1 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.0 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.1 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |